Journals
Evaluation of a Loop-Mediated Isothermal Amplification (LAMP) Method for the Detection of Salmonella spp. in Terms of Sensitivity and Applicability
A B S T R A C T
Salmonella spp. are important food-borne pathogens that can cause diseases in humans. Many detection methods have been established in Salmonella spp. using loop-mediated isothermal amplification (LAMP) or reverse transcription loop-mediated isothermal amplification (RT-LAMP). The detection limits of these assays varied from 1 CFU/reaction to 104 CFU/reaction, from 100 fg genomic DNA/reaction to 10 pg genomic DNA/reaction, or from 2.0×101 CFU/mL to 107 CFU/mL for food samples. In this study, LAMP assays were developed using genomic DNA for the detection of Salmonella spp. Two sets of LAMP primers were designed using the invA gene and the 16S-23S rRNA intergenic spacer region (ITS) of S. entericaas the target sequences for two LAMP assays. The detection limits of the two methods were respectively 20 pg S. entericaDNA/reaction and 10 pg S. entericaDNA/reaction at the optimized temperature, and the LAMP methods were of high repeatability and specificity for S. entericadetection. This study provides a baseline for the application of LAMP for the detection of food-borne pathogenic bacteria.
Keywords
Salmonella spp, loop-mediated isothermal amplification (LAMP), detection, sensitivity
Introduction
Food-borne diseases caused by pathogenic bacteria have become a major global public health issue, among which salmonellosis is the illness most often attributed to the consumption of poultry, beef, pork, eggs, milk, seafood, nut products, and fresh produce contaminated by Salmonella spp. [1, 2]. Salmonella spp. have been identified as the most frequent cause of food-borne infection outbreaks in many countries [3]. Therefore, rapid and sensitive methods for the detection of Salmonella spp. are needed to control and prevent salmonellosis outbreaks. Many methods, including culture-based, immunology-based, and nucleic acid- based, have been developed, among which more than twenty loop- mediated isothermal amplification (LAMP) or reverse transcription loop-mediated isothermal amplification (RT-LAMP) methods have been established targeting different specific genes of Salmonellaspp. [3].
Most of these LAMP methods used the invasion gene invA as the target sequence [4-13]. For example, Chen et al. developed specific LAMP methods utilizing the invE gene and 3 serotype-specific genes, fliC, lygD and STM4495 for the detection of Salmonella and 3 common Salmonella serotypes [14]. In the study of Tang et al., the LAMP assay with the fimY gene as the target sequence was established to detect Salmonella species in possibly infected ducks [15]. Okamura et al. developed the LAMP assays that amplified the rfbJ gene for detection of the O4 group of S. entericaand the insertion element of SE for detection of the O9 group of Salmonella in chickens [16, 17]. A sefA-based LAMP method was developed by Gong et al. for detection of S. enteritidis and S. gallinarum in chickens [18]. The phoP , hisJ, and rfbS genes of Salmonella spp. have also been used as target sequences for LAMP assays [2]. In spite of different genes as target sequences, all of these LAMP or RT-LAMP assays were of high specificity for Salmonella spp., but they varied greatly in sensitivity (detection limit).
When CFU/reaction, cfu/test, copies/reaction, and cells/test were used as the measurement units, the detection limits of 1 CFU per reaction, >2.2 cfu/test, 4 CFU/reaction, 6.0 cfu/test and 4.8 cfu/test, 15 copies/reaction, and 104 CFU/reaction, have been reported [4, 9, 12, 15, 18, 19]. With CFU/mL, CFU/25 ml, CFU/g and CFU/25g as the measurement units for detection of Salmonella spp. in food produces, the detection limits were determined to be 2.0×101 CFU/mL, 102 CFU/25g, 200 CFU/g, 103 CFU/ml, 104 CFU/25 ml, 104-106 CFU per 25 g, and 107 CFU/mL [8, 11-13, 17, 19].
When pg DNA/tube, fg DNA/tube, and pg/μL were used as the measurement units, the sensitivity of the LAMP assays was found to be 100 fg DNA/tube, 1 pg DNA/tube, and 10 pg/μL [3, 5, 7]. In conclusion, since the results of these LAMP assays in detection limits have been confusing to interpret, the LAMP technology has not been practically applied for the detection of Salmonella spp. In this study, genomic DNA from pure bacterial cultures of Salmonella spp. was used as template, LAMP assays with the invA gene and 16S-23S rRNA intergenic spacer region of S. entericaas the target sequences were developed, the specificity and sensitivity of LAMP assays were also evaluated.
Materials and Methods
I Primer Design for LAMP Assay
Two sets of LAMP primers targeting the invA gene (GenBank accession No. CP041208.1) and the 16S-23S ribosomal RNA intergenic spacer (ITS) (GenBank accession No. CP050716.1) of Salmonella entericaserotype Newport were designed using PrimerExplorer 5 (Link), and one set of primers was selected with Oligo 7 (Molecular Biology Insights, Inc. Colorado Springs, USA). The primer sequences are listed in (Table 1).
Table 1: LAMP primers targeting invA and ITS of Salmonella enterica.
|
Target (GenBank accession no.) |
Primer |
Sequence (5’–3’) |
|
invA (CP041208.1) |
F3 B3 FIP BIP LF |
GGAAAAAGAAGGGTCGTCGT ATGCTGTTATCGTCCAGGC CCGGCTCTTCGGCACAAGTAATTTTTGGACTGATTGGCGATCTCG AAGCTCAACTTGCGGAGCGTTTTTAACAATACTTCCGGCAGGC GGTACGGTCTCTGTAGAGACTTTAT |
|
16S-23S ribosomal RNA intergenic spacer CP050716.1 |
F3 B3 FIP BIP LF LB |
TCACACAGATTGTCTGATGA CGTGGAATAACGAAGCATAC TTATCAGGGGTGCGCTCTAATTTTAACGAGCAGTAAAACCTCTA GGTGAGGTCGGTGGTTCAAGTTTTTATGTGAGTTATTTCACAACGC CCACCTGAGCTACAAGCCTG AGGCCTACCAAATTTTCCCTGAAT |
II Bacteria Strains and Genomic DNA Extraction
The Salmonella entericaand non-Salmonella entericastrains used in this study are listed in (Table 2). All of the strains except the Listeria monocytogenes strains were cultured overnight at 37°C in Luria-Bertani (LB) broth, and the L. monocytogenes strains were cultured in DifcoTM Buffered Listeria Enrichment Broth Base (Becton, Dickinson and Company). Genomic DNA from the pure cultures was extracted using the DNeasy® Blood & Tissue Kit (QIAGEN N.V. Corporate), and the DNA templates were used to determine the sensitivity and specificity of the LAMP assays.
III Sensitivity Determination
The LAMP assay using the invA gene primers was performed in a 25 μL reaction mixture containing 0.8 µM each of forward inner primer (FIP) and backward inner primer (BIP), 0.2 µM each of forward outer primer (F3) and backward outer primer (B3), 0.4 µM of forward loop primer (LF), 1.4 mM dNTPs, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 6 mM MgSO4, 0.1% Triton X-100, 0.8 M betaine, 1×EvaGreen, 1×Rox, 8 U Bst 2.0 WarmStart DNA polymerase (New England Biolabs, Beverly, MA, USA), and serial dilutions ranging from 200 pg to 0.2 pg of Salmonella entericagenomic DNA [15]. The reaction mixtures were heated at the optimized temperature of 65°C for 60 min (30 s per cycle) using a StepOneTM System (Applied Biosystems, Foster City, CA, USA).
The LAMP assay with the 16S-23S ribosomal RNA intergenic spacer as the target sequence was modified according to the method of Wang et al. [20]. The reaction was performed in a 10 μL reaction mixture containing 0.8 µM each of forward inner primer (FIP) and backward inner primer (BIP), 0.2 µM each of forward outer primer (F3) and backward outer primer (B3), 0.4 µM of forward loop primer (LF) and backward loop primer (LB), 1.0 mM dNTPs, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 6 mM MgSO4, 0.1% Triton X-100, 7.5% DMSO, 1×EvaGreen, 1× Rox, 3.2 U Bst 2.0 WarmStart DNA polymerase (New England Biolabs, Beverly, Mass., USA.) and serial dilutions ranging from 10 pg to 0.01 pg of Salmonella entericagenomic DNA. The reaction mixture was heated at the optimized temperature 57°C for 60 min (30 s per cycle) using a StepOneTM System (Applied Biosystems, Foster City, CA, USA) [21].
IV Specificity Determination
The specificity of two LAMP assays with invA primers and ITS primers were determined using the bacterial strains listed in (Table 2), and the amount of DNA template used was 100 pg per reaction.
Results
I Detection Limit of the LAMP Assays
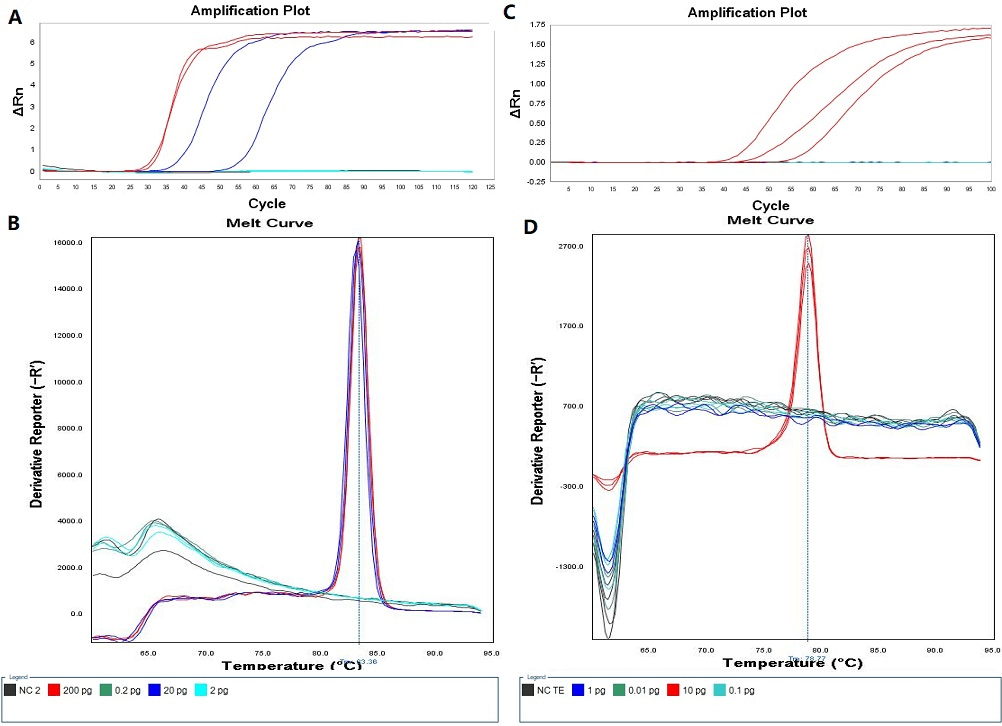
As shown in (Figure 1), the detection limit of the LAMP assay with invA primers was 20 pg per reaction, while the sensitivity of the LAMP assay with 16S-23S ribosomal RNA intergenic spacer as target sequence was 10 pg per reaction, and there was little significant difference in terms of detection limits for these two LAMP assays.
Table 2: Bacterial strains used in the study.
|
Bacterial Strain (Serotype) |
Bacterial Strain (Serotype) |
|
Escherichia coli O121:H19 |
Listeria monocytogenes J1-094 (1/2c) |
|
Escherichia coli O26:H11 |
Listeria monocytogenes C1-115 (3a) |
|
Escherichia coli O111:H8 |
Listeria monocytogenes J1-031 (4a) |
|
Escherichia coli O145:H2 |
Listeria monocytogenes W1-110 (4c) |
|
Escherichia coli O103:H2 |
Listeria monocytogenes ATCC19115 (4b) |
|
Escherichia coli O45:H12 |
Listeria innocua ATCC51742 |
|
Listeria monocytogenes J1-225 (4b) |
Listeria invanovii ATCC49954 |
|
Listeria monocytogenes J2-020 (1/2a) |
Salmonella Typhimuriam |
|
Listeria monocytogenes J2-064 (1/2b) |
Salmonella enterica serotype Newport |
|
Listeria monocytogenes J1-169 (3b) |
Escherichia coli O157:H7 933 |
|
Listeria monocytogenes J1-049 (3c) |
Escherichia coli O157:H7 B1409 |
|
Listeria monocytogenes M1-004 (N/A) |
|
|
Listeria monocytogenes M1-004 (N/A) |
|
All of the strains were from the culture collection of Eastern Regional Research Center (ERRC), Agricultural Research Service (ARS), United States Department of Agriculture (USDA).
Figure 1: Detection limits of two LAMP assays. A) Amplification plot of LAMP assay with invA primers. B) Melt curve of LAMP assay with invA primers. C) Amplification plot of LAMP assay with ITS primers. D) Melt curve of LAMP assay with ITS primers.
Figure 2: Specificities of the LAMP assay with invA primers and ITS primers. A) Amplification plot of the specificity determination for invA LAMP assay. B) Amplification plot of the specificity determination for ITS LAMP assay; non-Sa: 22 non-Salmonella entericastrains in (Table 2).
II Specificity of the LAMP Assay with ITS Primers
The specificities of the LAMP assay with ITS primers and the LAMP assays with invA primers were determined. As shown in (Figure 2), as for both LAMP assays, the six repeats of Salmonella entericaserotype Newport were positive, while the two repeats of 22 non-Salmonella entericastrains and two negative controls were negative; therefore, the LAMP assays were highly specific for Salmonella enterica.
Discussion
Two LAMP assays with the invA gene and 16S-23S rRNA intergenic spacer region of S. entericaas the target sequences were established in this study. The sensitivity of the assay with ITS primers (10 pg per reaction) was slightly higher than that of the assay with invA primers (20 pg per reaction), which is equivalent to 3×104 copies/reaction. The sensitivity was similar to that of the LAMP assays developed by Liu et al. and Zhang et al., but it was much lower than that of other reported LAMP assays in which there was up to four orders of magnitude difference [3-5, 9, 12, 14, 15, 18, 19].
One important factor for differences in sensitivity is that the DNA preparation methods can have a great effect on the LAMP reaction. When the DNA template was prepared by a boiling method, the detection limit was as low as 1 CFU per reaction; it was proposed that the boiling and cooling process provided the single-strand template for the LAMP reaction [4, 5, 10, 12-15]. A commercial DNA preparation kit was used by Liu et al., and in our study, the prepared DNA template was double stranded, and the sensitivity was as low as 10 pg per reaction [3]. The mechanisms of Polymerase Chain Reaction (PCR), Helicase-dependent Amplification (HDA) and Recombinase Polymerase Amplification (RPA) assays for amplification of double stranded DNA are well-established; however, there is so far no report on the mechanism of the LAMP reaction with double stranded DNA as template, which warrants further study.
Other factors related to sensitivity include the method used to determine results, and use of enrichment, reverse transcription, etc. It was reported by Zhang et al. that the reduction of the detection limit, from 104 copies/reaction to 102 copies/reaction, could be the result of using visual determination by fluorescence instead of turbidity [9]. The LAMP assays could detect 104 CFU/25 ml without enrichment and 100 CFU/25 ml with an 8 h enrichment for Salmonella spp., Techathuvanan et al. improved the Salmonella detection limit from 106 CFU/25 g to 102 CFU/25 g for both pork chop and sausage samples using a 10 h enrichment [8, 12]. With detection of Salmonella spp. with RT-LAMP, the 6 h enrichment made little difference and only improved the detection limits from 107 CFU/25 mL to 106 CFU/25 mL [11]. It could be that competitive or non-competitive inhibitors existed between reactions catalyzed by reverse transcriptase and Bst DNA polymerase (Large Fragment), which also needs further study.
The LAMP assays with the invA gene and the 16S-23S ribosomal RNA intergenic spacer as the target sequence were established in this study. Like most previous reports, they were highly specific to Salmonella enterica[2]. It was also proven by Wang et al. that the presence of high concentrations of non-target genomic DNA neither adversely affected the amplification efficiency nor generated significant background [5]. In conclusion, the LAMP assay can be used to detect Salmonella spp. in different food matrices sensitively and selectively, given that a proper DNA preparation method was used after enrichment.
Acknowledgment
This study was funded by the National Key Research and Development Program of China (2016YFD0500704-4) and Henan Science and Technology Plan Project (182102110285).
Conflicts of Interest
None.
Article Info
Article Type
Research ArticlePublication history
Received: Sat 06, Jun 2020Accepted: Fri 19, Jun 2020
Published: Fri 03, Jul 2020
Copyright
© 2023 Yanhong Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.JFNM.2020.02.03
Author Info
Deguo Wang Meng Zhang Yanhong Liu Yongzhen Wang
Corresponding Author
Yanhong LiuMolecular Characterization of Foodborne Pathogens Research Unit, Eastern Regional Research Center, Agricultural Research Service, United States Department of Agriculture, Wyndmoor, Pennsylvania, USA
Figures & Tables
Table 1: LAMP primers targeting invA and ITS of Salmonella enterica.
|
Target (GenBank accession no.) |
Primer |
Sequence (5’–3’) |
|
invA (CP041208.1) |
F3 B3 FIP BIP LF |
GGAAAAAGAAGGGTCGTCGT ATGCTGTTATCGTCCAGGC CCGGCTCTTCGGCACAAGTAATTTTTGGACTGATTGGCGATCTCG AAGCTCAACTTGCGGAGCGTTTTTAACAATACTTCCGGCAGGC GGTACGGTCTCTGTAGAGACTTTAT |
|
16S-23S ribosomal RNA intergenic spacer CP050716.1 |
F3 B3 FIP BIP LF LB |
TCACACAGATTGTCTGATGA CGTGGAATAACGAAGCATAC TTATCAGGGGTGCGCTCTAATTTTAACGAGCAGTAAAACCTCTA GGTGAGGTCGGTGGTTCAAGTTTTTATGTGAGTTATTTCACAACGC CCACCTGAGCTACAAGCCTG AGGCCTACCAAATTTTCCCTGAAT |
Table 2: Bacterial strains used in the study.
|
Bacterial Strain (Serotype) |
Bacterial Strain (Serotype) |
|
Escherichia coli O121:H19 |
Listeria monocytogenes J1-094 (1/2c) |
|
Escherichia coli O26:H11 |
Listeria monocytogenes C1-115 (3a) |
|
Escherichia coli O111:H8 |
Listeria monocytogenes J1-031 (4a) |
|
Escherichia coli O145:H2 |
Listeria monocytogenes W1-110 (4c) |
|
Escherichia coli O103:H2 |
Listeria monocytogenes ATCC19115 (4b) |
|
Escherichia coli O45:H12 |
Listeria innocua ATCC51742 |
|
Listeria monocytogenes J1-225 (4b) |
Listeria invanovii ATCC49954 |
|
Listeria monocytogenes J2-020 (1/2a) |
Salmonella Typhimuriam |
|
Listeria monocytogenes J2-064 (1/2b) |
Salmonella enterica serotype Newport |
|
Listeria monocytogenes J1-169 (3b) |
Escherichia coli O157:H7 933 |
|
Listeria monocytogenes J1-049 (3c) |
Escherichia coli O157:H7 B1409 |
|
Listeria monocytogenes M1-004 (N/A) |
|
|
Listeria monocytogenes M1-004 (N/A) |
|
All of the strains were from the culture collection of Eastern Regional Research Center (ERRC), Agricultural Research Service (ARS), United States Department of Agriculture (USDA).

References
- Ohtsuka K, Yanagawa K, Takatori K, Hara Kudo Y (2005) Detection of Salmonella enterica in Naturally Contaminated Liquid Eggs by Loop-Mediated Isothermal Amplification, and Characterization of Salmonella Isolates. Appl Environ Microbiol 71: 6730-6735. [Crossref]
- Kokkinos PA, Ziros PG, Bellou M, Vantarakis A (2014) Loop-Mediated Isothermal Amplification (LAMP) for the Detection of Salmonella in Food. Food Anal Method 7: 512-526.
- Liu N, Zou D, Dong D, Yang Z, Ao D et al. (2017) Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci Rep 7: 45601. [Crossref]
- Hara Kudo Y, Yoshino M, Kojima T, Ikedo M (2005) Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett 253: 155-161. [Crossref]
- Wang L, Shi L, Alam MJ, Geng Y, Li L (2008) Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res Int 41: 69-74.
- Lu Y, Yang W, Shi L, Li L (2009) Specific Detection of Viable Salmonella Cells by an Ethidium Monoazide-Loop Mediated Isothermal Amplification (EMA-LAMP) Method. J Health Sci 55: 820-824.
- Zhao X, Wang L, Chu J, Li Y, Li Y et al. (2010) Development and application of a rapid and simple loop-mediated isothermal amplification method for food-borne Salmonella detection. Food Sci Biotechnol 19: 1655-1659.
- Techathuvanan C, Draughon FA, D'Souza DH (2010) Loop-Mediated Isothermal Amplification (LAMP) for the Rapid and Sensitive Detection of Salmonella Typhimurium from Pork. J Food Sci 75: M165- M172. [Crossref]
- Zhang G, Brown EW, Gonzalez Escalona N (2011) Comparison of Real-Time PCR, Reverse Transcriptase Real-Time PCR, Loop-Mediated Isothermal Amplification, and the FDA Conventional Microbiological Method for the Detection of Salmonella spp. in Produce. Appl Environ Microbiol 77: 6495-6501. [Crossref]
- Shao Y, Zhu S, Jin C, Chen F (2011) Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int J Food Microbiol 148: 75-79. [Crossref]
- Techathuvanan C, D’Souza DH (2012) Reverse-Transcriptase Loop-Mediated Isothermal Amplification as a Rapid Screening/Monitoring Tool for Salmonella Enterica Detection in Liquid Whole Eggs. J Food Sci 77: M200-M205. [Crossref]
- Yang Q, Chen S, Ge B (2013) Detecting Salmonella Serovars in Shell Eggs by Loop-Mediated Isothermal Amplification. J Food Prot 76: 1790-1796. [Crossref]
- Yang Q, Wang F, Jones KL, Meng J, Prinyawiwatkul W et al. (2015) Evaluation of loop-mediated isothermal amplification for the rapid, reliable, and robust detection of Salmonella in produce. Food Microbiol 46: 485-493. [Crossref]
- Chen Z, Zhang K, Yin H, Li Q, Wang L et al. (2015) Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification Method. Food Sci Human Wellness 4: 75-79.
- Tang T, Cheng A, Wang M, Li X, He Q et al. (2012) Development and clinical verification of a loop-mediated isothermal amplification method for detection of Salmonella species in suspect infected ducks. Poultry Sci 91: 979-986. [Crossref]
- Okamura M, Ohba Y, Kikuchi S, Takehara K, Ikedo M et al. (2009) Rapid, Sensitive, and Specific Detection of the O4 Group of Salmonella enterica by Loop-Mediated Isothermal Amplification. Avian Dis 53: 216-221. [Crossref]
- Okamura M, Ohba Y, Kikuchi S, Suzuki A, Tachizaki H et al. (2008) Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens. Vet Microbiol 132: 197-204. [Crossref]
- Gong J, Zhuang L, Zhu C, Shi S, Zhang D et al. (2016) Loop-Mediated Isothermal Amplification of the sefA Gene for Rapid Detection of Salmonella Enteritidis and Salmonella Gallinarum in Chickens. Foodborne Pathog Dis 13: 177-181. [Crossref]
- Fenxia F, Pengcheng D, Biao K, Yan M (2015) The Development and Evaluation of a Loop-Mediated Isothermal Amplification Method for the Rapid Detection of Salmonella enterica serovar Typhi. Plos One 10: e0124507. [Crossref]
- Wang D, Brewster JD, Paul M, Tomasula PM (2015) Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification. Molecules 20: 6048-6059. [Crossref]
- Wang D, Wang Y, Zhu K, Shi L, Zhang M et al. (2019) Detection of Cassava Component in Sweet Potato Noodles by Real-Time Loop-mediated Isothermal Amplification (Real-time LAMP) Method. Molecules 24: 2043. [Crossref]
