Antibacterial Activity of Extracts of Coprinopsis Cinerea, A Coprophilous Fungus Against Multidrug Resistant Nosocomial Pathogens
Antibacterial Activity of Extracts of Coprinopsis Cinerea, A Coprophilous Fungus Against Multidrug Resistant Nosocomial Pathogens
A B S T R A C T
Our study explored Coprinopsis cinerea (C2), a coprophilous basidiomycetous fungus isolated from horse dung to combat beta-lactamase and carbapenamase producing multidrug resistant nosocomial pathogens in vitro. The isolated strain was cultivated under sub-merged fermentation (SmF) and solid-state fermentation (SSF) for 10 days at 30°C and at pH 6 in dark. After the growth period, the extracellular metabolites were extracted using polar and non-polar solvents and the extracts were subjected for antibacterial activity by agar well diffusion, microbroth dilution and time-kill kinetics assay. The methanolic extract of fruit bodies and mycelial biomass of C. cinerea (C2) grown under wheat flour agar and wheat flour broth respectively, showed significant antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and carbapenamase producing Klebsiella pneumoniae with a zone of inhibition ranging between 10 and 14 mm in diameter. On the contrary, ethyl acetate seems to be the effective solvent for extraction of antimicrobial compounds from culture filtrate of Smf and SSF grown C. cinerea (C2). Among the solid substrates (agrowaste materials), wheat bran supported maximum growth and antimicrobial metabolite production with a significant zone of inhibition ranging between 20 and 22 mm. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of ethyl acetate extract of C. cinerea (C2) culture filtrate was 3.12-12.5 mg/mL and 6.25-25 mg/mL, respectively. The time-kill kinetics assay showed the extracted metabolites of culture filtrate was bactericidal in nature.
Keywords
Coprinopsis cinerea, coprophilous, nosocomial, β-lactamase, carbapenamase, time-kill kinetics
Introduction
Drugs derived from the natural sources play a major role in the prevention and treatment of human diseases caused by multi-drug resistant pathogens [1]. Therefore, there is a constant search for newer resources for the isolation of antimicrobial compounds, one such example is mushrooms. Mushrooms are rapidly growing, often wood colonizing fleshy fungi, belonging to Basidiomycetes of a division Eumycota and characterized by the formation of basidiospores. Many secondary or specific metabolites produced in the life cycle of mushrooms have been discovered and demonstrated to have antimicrobial, antioxidant, and anti-inflammatory, in addition to their nutritional and culinary properties [2]. Mushrooms have developed and adapted their metabolism in their habitat for survival and for competing with other organisms. Many wild mushrooms and coprophilous mushrooms have been considered to have antimicrobial properties and are more often used as bioresources for identifying antimicrobials than cultivated mushrooms [2, 3].
Although many varieties of mushrooms have been discovered, only 2000 among 140000 identified species are safe as food and only 158 species from 88 genera have been recognized to have antimicrobial properties [4]. Coprinopsis cinerea is a coprophilous basidiomycetous mushroom, which in nature grows on dung [5, 6]. Coprinopsis cinerea can readily fructify under laboratory conditions and thus serves as a model basidiomycete for fruiting body development [7]. Several reports are available on the application of Coprinopsis cinerea in the field of genetics but only few reports are available on the evaluation of antimicrobial activity on this mushroom. Therefore, the present study was aimed to evaluate the antibacterial compounds produced during the growth period of Coprinopsis cinerea against multi-drug resistant nosocomial pathogens.
Materials & methods
I Chemicals
The chemicals and reagents (analytical grade) used in the media and reagent preparation were purchased from Sisco Research Laboratory (SRL) and Himedia, Mumbai, India.
II Bacterial pathogens
Nine clinical isolates, Staphylococcus aureus (3 strains), Klebsiella pneumoniae (3 strains), E. coli (2 strains), and Pseudomonas aeruginosa (1 strain) were procured from Mahaveer Jain hospital, Bangalore, India. Staphlococcus aureus (MTCC 3160) and Klebsiella pneumoniae (MTCC 432) were procured from Microbial type culture collection (MTCC) centre, Chandigarh, India. All the cultures were maintained in nutrient agar at 4°C until use.
III Mushroom culture
The pure culture of Coprinopsis cinerea (C2) (KX468975) isolated by us from horse dung was used for the present study and it was maintained in 2% wheat flour agar (WFA) medium at 4°C until use [8].
IV Growth of Coprinopsis cinerea
Two methods of growth such as submerged and solid substrate fermentation was carried out [9, 10].
i. Submerged fermentation
Submerged fermentation was carried out according to the method described by Yamac and Bilgili with little modification [9]. Two hundred ml of 2% wheat flour broth adjusted to pH 7.0 was taken in 500ml Erlenmeyer flasks, sterilized and inoculated with 5mm C. cinerea mycelial agar plugs. The inoculated flasks were shaken at 120 rpm at 30º C in dark. The culture broth was separated from mycelial biomass by filtration using Whatman no.1 paper after 10 days of growth period.
ii. Solid state fermentation
Wheat bran, wheat straw, paddy straw and rice bran were used as substrates for the cultivation of C. cinerea (C2) under solid substrate fermentation [9]. Forty grams of each substrate was taken in 500ml Erlenmeyer flasks and were moistened with 70% distilled water. After sterilization the flasks were inoculated with 5mm mycelia agar plugs from the wheat flour agar (WFA) culture. The inoculated flasks were incubated at 30º C for 10days in dark.
V Extraction of metabolites from mycelial biomass and fruit bodies
Extraction of metabolites from mycelial biomass and fruit bodies was carried out according to Yamac and Bilgili [9]. The ten days grown mycelial biomass and fruit bodies of Coprinopsis cinerea (C2) were dried in an oven at 50º C until constant weight was obtained. Ten grams of dried material of each was extracted with 100 mL of analytical grade methanol (Himedia, India) for 72 hrs, the process was continued three times. The combined extracts were filtered through Whatman filter paper 1 and concentrated to dryness using rotary vacuum evaporator.
VI Extraction of metabolites from culture filtrate
Extraction of metabolites from Coprinopsis cinerea (C2) culture filtrate was carried out according to Yamac and Bilgili with modification [9]. Hundred milliliter of culture broth obtained after submerged fermentation was extracted three times with 100mL of hexane, chloroform and ethyl acetate separately. All the solvent fractions were concentrated under vacuum at 40º C to dryness using Rotavap (Heidolph, Germany). The dried extract was dissolved in DMSO (100mg/mL) and stored at -20ºC until use.
VII Extraction of metabolites from solid state fermented culture
Extraction of metabolites from solid state fermented culture was carried out according to Machado et al. with modification [10]. At the end of fermentation process, the fermented matter in the whole flask was mixed with 160mL of ethyl acetate and shaken in an orbital shaker at room temperature for 24 hrs. The extraction process was conducted thrice with the same volume of solvents; the combined extract was concentrated to dryness at 40º C using rotary vacuum evaporator (Heidolph, Germany) and dissolved in DMSO (100mg/mL) and stored at -20ºC until use.
VIII Characterization of antibiotic resistance and sensitivity profile of pathogens
Characterization of antibiotic resistance and sensitivity profile of bacterial pathogens were performed using VITEK 2 Compact card (Biomerieux, Lyon, France). These cards allow the simultaneous determination of susceptibility to antimicrobial agents and the strain identification, including aerobic and facultative anaerobic Gram-negative bacilli (VITEK 2 Compact AST-P619). The sensitivity of the micro-organisms to each antibiotic was identified by MIC.
IX Antibacterial activity by agar well diffusion assay
For evaluation of antibacterial activities of the extracts agar well diffusion method was used as described by Appiah et al. with modification [11]. Twenty mL of sterile Mueller-Hinton agar was poured into petriplates. The bacterial inoculum concentration was adjusted to 0.5 McFarland standard (1.5×108 CFU/mL) and inoculated separately by swabbing along the media. Four wells (5mm) equidistant from each other were created with a sterile cork borer. The wells were filled with 100 µL of 100mg/mL of extracts. The plates were kept in a refrigerator for 2 hrs to allow diffusion of the extracts. The zones of growth inhibition were measured after 24 hrs of incubation at 37°C. The procedure was performed in triplicates and the mean zones of growth inhibition were determined.
X Determination of minimum inhibitory concentration (MIC)
Minimum inhibitory concentration of C. cinerea (C2) extracts was determined based on a broth micro dilution method in 96 well micro-titre plates according to Yamac and Bilgili with modification. Bacterial strains were cultured overnight at 37ºC on Mueller Hinton (MH) broth and adjusted to final density of 108 CFU/mL by 0.5 McFarland standards [9]. The fungal extract (100mg/mL) was dissolved in DMSO and stored at -20°C until use. Mueller Hinton Broth (MHB) was prepared and sterilized using autoclave. Ninety micro litres (90µl) of sterile MHB was dispensed into each well using a sterile pipette. Hundred micro litres (100µl) of crude extract (25mg/mL) was added to first well and was serially diluted two fold till the sixth well and final mixture was discarded. Further, 100µl of tetracycline (500µg/mL) and DMSO were added to seventh and eighth well and considered as positive and negative control respectively. Each well was inoculated with 10µL of overnight grown bacterial cultures. The concentration of the extract was adjusted to give final concentration of 50, 25, 12.5, 6.25, 3.125, 1.563mg/mL and incubated at 37°C for 24 h. An hour before the end of incubation period 40μl of 0.2mg/ml INT (p-iodonitrotetrazolium violet) solution was added to each well and colour development was observed till 3hrs. The lowest concentration where growth is inhibited was recorded as the minimum inhibitory concentration (MIC). This was indicated by a well with a decreased colour or no colour after incubation with INT (p-iodonitrotetrazolium violet).
XI Determination of minimum bactericidal concentration (MBC)
A sample was taken from each well that exhibited no bacterial growth from 96-well plates and was streaked onto freshly prepared Mueller Hinton agar plates. The plates were then incubated at 37ºC for 24hrs. After the incubation, the plates were observed for growth of the bacteria. The MBC was determined to be the concentration at which no bacterial growth was observed.
XII Time-Kill kinetic assay
Time-kill kinetics studies were done according to Jeyanthi and Velusamy [12]. The 0.5,1 and 2×MIC of ethyl acetate extract was added separately to the bacterial culture density of 108CFU/mL and incubated at 37°C. Bacterial culture without the extract was used as growth control. At 2 h time intervals for a period of 24 h, the samples were withdrawn, serially diluted, plated on to Muller-Hinton agar (MHA) medium and incubated at 37°C for 24 h. Rates of killing were determined by measuring the reduction in viable bacteria (log10 CFU/mL). The killing curve was constructed by plotting number of viable cells against time.
XIII Statistical analysis
All the data were expressed as mean±Standard Deviation. The differences between the means were analysed by Tukey’s test of one way ANOVA using SPSS 20 version (SPSS Inc., Chicago., IL, USA).
Table 1: Antibiotic susceptibility profile of Gram-positive bacteria (MIC values in µg/ml)
|
Antibiotic |
Staphylococcus aureus (SA 1) |
Staphylococcus aureus (SA 2) |
Staphylococcus aureus (SA 3) |
|||
|
MIC |
INT |
MIC |
INT |
MIC |
INT |
|
|
Benzylpenicillin |
> = 0.5 |
R |
> = 0.5 |
R |
> = 0.5 |
R |
|
Cefoxitin screen |
POS |
+ |
POS |
+ |
POS |
+ |
|
Ciprofloxacin |
>=18 |
R |
>=8 |
R |
>=8 |
R |
|
Clindamycin |
0.25 |
S |
0.25 |
S |
>=4 |
R |
|
Daptomycin |
0.25 |
S |
0.5 |
S |
0.5 |
S |
|
Erythromycin |
1 |
S |
<=0.25 |
S |
>=8 |
R |
|
Gentamicin |
>=16 |
R |
4 |
S |
< = 0.5 |
S |
|
Inducible Clindamycin resistance |
NEG |
- |
NEG |
- |
NEG |
- |
|
Levofloxacin |
4 |
I |
4 |
I |
4 |
1 |
|
Linozolid |
2 |
S |
2 |
S |
2 |
S |
|
Nitrofurantoin |
32 |
S |
< = 16 |
S |
32 |
S |
|
Oxacillin |
> = 4 |
R |
> = 4 |
R |
> = 4 |
R |
|
Rifampicin |
< = 0.03 |
S |
< = 0.03 |
S |
32 |
S |
|
Teicoplanin |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
|
Tetracyclin |
<=1 |
S |
<=1 |
S |
<=1 |
S |
|
Tigecycline |
< = 0.12 |
S |
< = 0.12 |
S |
< = 0.12 |
S |
|
Trimethoprim/Sulfamethoxazole |
>=320 |
R |
80 |
R |
>=320 |
R |
|
Vancomycin |
1 |
S |
< = 0.5 |
S |
1 |
S |
S, Susceptible; I, intermediate; R, resistant (this classification was made according to the interpretative breakpoints suggested by Clinical and Laboratory Standards Institute – CLSI); POS, positive to Cefoxitin screening; NEG, negative to Cefoxitin screening, MIC, Minimum inhibitory concentration, INT, Interpretation
Results & Discussion
The growing numbers of antimicrobial-resistant pathogens, which are increasingly associated with nosocomial infection, place a significant burden on healthcare systems and have important global economic costs [13]. Therefore, it is important to find novel antimicrobials effective against those multi-drug resistant pathogens. In our study, the clinical isolates obtained from Mahaveer Jain Hospital, Bangalore were subjected for evaluation of antibacterial activity of Coprinopsis cinerea (C2) extracts. (Table 1 and 2) shows the antibiotic resistance and sensitivity profile of clinical isolates used in the present work. Based on the interpretation of the results we found that the isolates are multi-drug resistant bacteria, which are resistant to more than three antibiotics. Klebsiella pneumoniae and Pseudomonas aeruginosa are the Gram-negative bacteria with the highest number of resistance, especially to beta-lactam antibiotics. E. coli showed resistance to few beta-lactam antibiotics but more sensitive to macrolides. All the Staphylococcus aureus strains used in the study were resistant to beta-lactams, ciproplaxacin, oxacillin and trimethoprin and more sensitive to linozolid and macrolides antibiotics.
Classic solvent extraction is the most widely used method for the extraction of active compounds from natural sources. The solvent used for extraction has a profound effect on the quality, quantity, and safety of the recovered products. Therefore, selection of the right solvent for maximum extraction of metabolites of our interest is essential. Antimicrobial activity of the different solvent extracts of Coprinopsis cinerea (C2) against multi-drug resistant bacteria is shown in (Figure 1, 2, 3, and 4). It should be pointed out that the majority of the extracts did not show antibacterial activity against E. coli and Pseudomonas aeruginosa which is in agreement with the results previously reported by Alves et al. even when higher concentrations were used [1]. However, a study describes the methanolic extracts of Polyporus giganteus having high antibacterial activity against E. coli [14]. Methanol is the most commonly used solvents in extracting antimicrobial components of microbes [15]. It can be used for extracting small-molecular weight phenolic compounds and certain other compounds with strong polarity, such as saccharides and polysaccharide-binding proteins.
Table 2: Antibiotic susceptibility profile of Gram-negative bacteria (MIC values in µg/ml)
|
Antibiotic |
E. coli (EC 1) |
E. coli (EC 2) |
Klebsiella pneumoniae (KP1) |
Klebsiella pneumoniae (KP2) |
Klebsiella pneumoniae (KP3) |
Pseudomonas aeruginosa |
||||||
|
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
|
|
Amikacin |
< = 2 |
S |
< = 2 |
S |
>=64 |
R |
< = 2 |
S |
>=64 |
R |
>=64 |
R |
|
Amoxicillin/Clavulanic acid |
>=32 |
R |
4 |
S |
>=32 |
R |
>=32 |
R |
>=32 |
R |
na |
|
|
Ampicillin |
>=32 |
R |
>=32 |
R |
>=32 |
R |
>=32 |
R |
>=32 |
R |
na |
|
|
Cefepime |
2 |
S |
16 |
R |
>=64 |
R |
2 |
S |
>=64 |
R |
>=64 |
R |
|
Cefoperazone/Sulbactam |
< = 8 |
S |
16 |
S |
>=64 |
R |
32 |
I |
>=64 |
R |
>=64 |
R |
|
Ceftazidime |
na |
na |
na |
na |
na |
>=64 |
R |
|||||
|
Ceftriaxone |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Cefuroxime |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Cefuroxime Axetil |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Ciprofloxacin |
>=4 |
R |
0.5 |
S |
>=4 |
R |
>=4 |
R |
>=4 |
R |
>=4 |
R |
|
Colistin |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
|
Doripenem |
na |
na |
na |
na |
na |
>=8 |
R |
|||||
|
Ertapenem |
< = 0.5 |
S |
< = 0.5 |
S |
>=8 |
R |
< = 0.5 |
S |
>=8 |
R |
na |
|
|
Gentamicin |
< = 1 |
S |
< = 1 |
S |
4 |
S |
>=16 |
R |
< = 1 |
S |
>=16 |
R |
|
Imipenem |
< = 0.25 |
S |
< = 0.25 |
S |
2 |
R |
< = 0.25 |
S |
2 |
R |
>=16 |
R |
|
Levofloxacin |
na |
na |
na |
na |
na |
>=8 |
R |
|||||
|
Meropenem |
< = 0.25 |
S |
< = 0.25 |
S |
>=16 |
R |
< = 0.25 |
S |
>=16 |
R |
>=16 |
R |
|
Minocycline |
na |
na |
na |
na |
na |
>=16 |
R |
|||||
|
Nalidixic acid |
> = 32 |
R |
na |
> = 32 |
R |
na |
na |
na |
||||
|
Nitrofurantoin |
< = 16 |
S |
na |
256 |
R |
na |
na |
na |
||||
|
Piperacillin/Tazobactam |
< = 4 |
S |
8 |
S |
>=128 |
R |
>=128 |
R |
>=128 |
R |
>=128 |
R |
|
Tigecycline |
< = O.5 |
S |
< = O.5 |
S |
> = 8 |
R |
< = O.5 |
S |
4 |
I |
>=8 |
R |
|
Tricarcillin/Clavulanic acid |
na |
na |
na |
na |
na |
>=128 |
R |
|||||
|
Trimethoprim/Sulfamethoxazole |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
interpretative breakpoints suggested by Clinical and Laboratory Standards Institute – CLSI); na, not applicable; POS, positive to Cefoxitin screening; NEG, negative to Cefoxitin screening, MIC, Minimum inhibitory concentration, INT, Interpretation
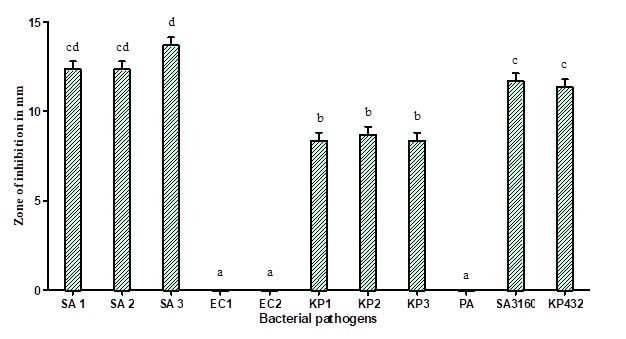
Figure 1 : Antibacterial activity of methanolic extracts of fruit bodies of Coprinopsis cinerea (C2): SA 1 (Methicillin-resistant Staphylococcus aureus), SA 2 (Methicillin-resistant Staphylococcus aureus), SA 3 (Methicillin-resistant Staphylococcus aureus), KP 1 (Klebsiella pneumoniae), KP 2 (Klebsiella pneumoniae), KP 3 (Klebsiella pneumoniae), SA 3160 (Staphylococcus aureus MTCC 3160), and KP 432 (Klebsiella pneumoniae MTCC 432). The results are expressed in means±SD (n=3), the same letters in the bar do not differ significantly as per Tukey’s test (P ≤ 0.05).
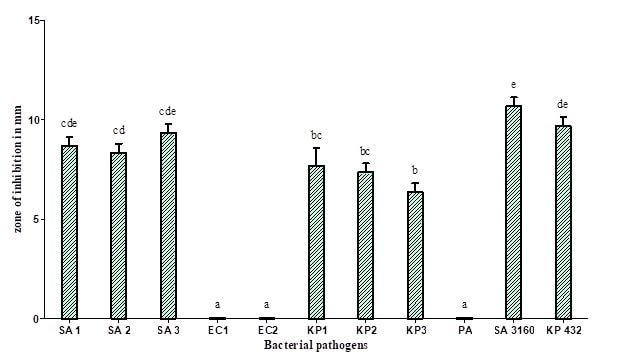
Figure 2 : Antibacterial activity of methanolic extract of mycelia of Coprinopsis cinerea (C2): SA 1 (Methicillin-resistant Staphylococcus aureus), SA 2 (Methicillin-resistant Staphylococcus aureus), SA 3 (Methicillin-resistant Staphylococcus aureus), KP 1 (Klebsiella pneumoniae), KP 2 (Klebsiella pneumoniae), KP 3 (Klebsiella pneumoniae), SA 3160 (Staphylococcus aureus MTCC 3160), and KP 432 (Klebsiella pneumoniae MTCC 432). The results are expressed in means±SD (n=3), the same letters in the bar do not differ significantly as per Tukey’s test (P ≤ 0.05).
In the present study, the methanolic extract of fruit bodies and mycelium of Coprinopsis cinerea (C2) showed significant antibacterial activity against all the strains of methicillin-resistant Staphylococcus aureus and carbapenamase positive Klebsiella pneumoniae with a zone of inhibition of growth ranging between 9 and 14 mm in diameter (Figure 1 and 2). The methanolic extracts of C. cinerea (C2) fruit bodies and mycelial biomass showed strong antibacterial activity against Gram-positive bacteria compared to Gram-negative bacteria. The result is much similar to the work reported by Oyetayo, who studied the antibacterial activity of extracts of Lentinus subnudus against Bacillus cereus, Staphylococcus aureus and Salmonella typhimurium, showing similar such results of ours [16]. Venturini et al. reported that the aqueous and methanolic extracts of vegetative mycelium and fruiting bodies of Lentinus edodes, Craterellus cornucopioides, Tricholoma equestre contains substances against Listeria monocytogens, Clostridium perfringens and Bacillus cereus [17]. In general, all types of mushrooms produce promising metabolites possessing antimicrobial activity, Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis are the most susceptible bacteria in general. Methanolic extracts of Agaricus bisporus, Boletus edulis, Cantharellus cibarius, Clitocybe alexandri, Lepista nuda, Coriolus versicolor, Pleurotus ostreatus and different Lactarius sp. demonstrated good inhibitory activity against Gram-positive bacteria [1, 18-22].
Our study demonstrated that ethyl acetate is the most suitable solvent for the extraction of metabolites released by C. cinerea (C2) grown under SmF and SSF. The extract showed significant antibacterial activity against Staphylococcus aureus and Klebsiella pneumoniae with a zone of inhibition of growth ranging between 11 and 14 mm to 18 and 11mm in diameter, compared to other solvents used (Figure 3). Our results are in agreement with the results of Venturini et al. who reported Hydnum repandum and H. rufescens showed significant antibacterial activity against Staphylococcus aureus and Clostridium perfringens [17]. The effective solvents reported by them were hexane and ethyl acetate.
Figure 3: Antibacterial activities of extracts of culture filtrate of Coprinopsis cinerea (C2): SA1 (Methicillin-resistant Staphylococcus aureus), SA2 (Methicillin-resistant Staphylococcus aureus), SA3 (Methicillin-resistant Staphylococcus aureus), KP1 (Klebsiella pneumoniae), KP2 (Klebsiella pneumoniae), KP 3 (Klebsiella pneumoniae), SA 3160 (Staphylococcus aureus MTCC 3160), and KP 432 (Klebsiella pneumoniae MTCC 432).
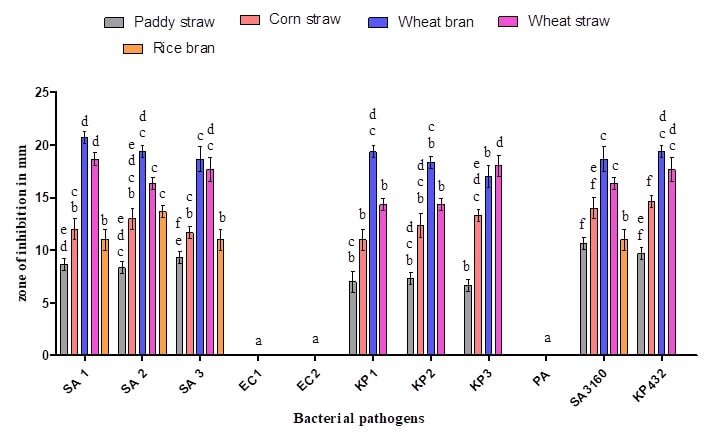
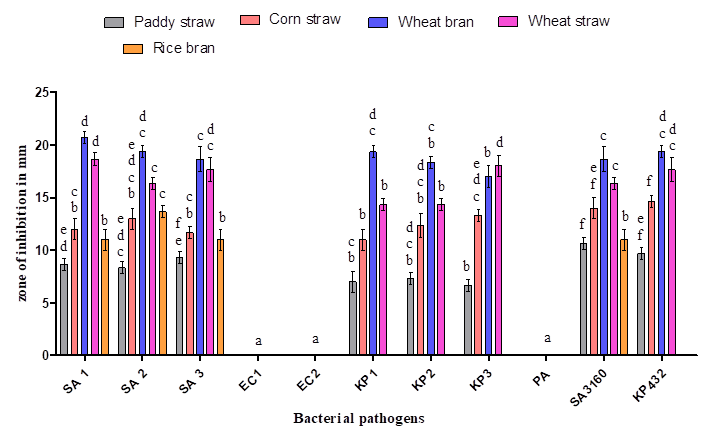
Figure 4: Antibacterial activity of ethyl acetate extract of Coprinopsis cinerea (C2) grown on various solid substrates: SA 1 (Methicillin-resistant Staphylococcus aureus), SA 2 (Methicillin resistant Staphylococcus aureus), SA 3 (Methicillin-resistant Staphylococcus aureus), KP 1 (Klebsiella pneumoniae), KP 2 (Klebsiella pneumoniae), KP 3 (Klebsiella pneumoniae), SA 3160 (Staphylococcus aureus MTCC 3160), and KP 432 (Klebsiella pneumoniae MTCC 432). The results are expressed in means±SD (n=3), the same letters in the bar do not differ significantly as per Tukey’s test (P ≤ 0.05).
Table 3: Minimum inhibitory concentration (MIC) and Minimum bactericidal concentration (MBC) of ethyl acetate extract of C. cinerea (C2) against multidrug resistant bacteria (mg/ml)
|
Name of the pathogen |
Solid substrate |
|
|
|
Wheat bran |
|
|
|
MIC |
MBC |
|
Staphylococcus aureus (SA 1) |
3.12 |
6.25 |
|
Staphylococcus aureus (SA 2) |
6.25 |
12.5 |
|
Staphylococcus aureus (SA 3) |
6.25 |
6.25 |
|
Klebsiella pneumoniae (KP1) |
3.12 |
6.25 |
|
Klebsiella pneumoniae (KP2) |
3.12 |
6.25 |
|
Klebsiella pneumoniae (KP3) |
6.25 |
12.5 |
|
Klebsiella pneumoniae (432) |
6.25 |
12.5 |
|
Staphylococcus aureus (3160) |
3.12 |
6.25 |
Five agro-waste substrates (Wheat straw, wheat bran, paddy straw, rice bran and corn straw) were used in the present study for the cultivation of C. cinerea (C2). Among which, wheat bran yielded significant amount of antibacterial compounds by supporting growth of the fungus and showed maximum activity against Staphylococcus aureus and Klebsiella pneumoniae with the zone of inhibition ranging between 18 and 22mm in diameter compared to other agro-waste substrates used in the present study (Figure 4). In the literature, usage of wheat bran as a solid substrate for the production of antibiotics such as rifamycin B, rifamycin SV, cephalosporins, meroparamycin etc using filamentous fungi is well documented [23, 24, 25]. The minimum inhibitory concentration (MIC) of ethyl acetate extract of C. cinerea (C2) culture filtrate was found to be 3.12 mg/mL for Staphylococcus aureus and 6.25 mg/ml for Klebsiella pneumoniae (Table 3), it is little higher to the findings of Taofiq et al. (MIC = 2.5 mg/mL) [26]. Similarly, the minimum bactericidal concentration (MBC) was found to be 6.25 mg/ml for Staphylococcus aureus and 12.5 mg/ml for Klebsiella pneumoniae (Table 3). Nedelkoska et al. obtained MIC and MBC ranging from 5 to 50 mg/mL while evaluating the antibacterial activity of five mushrooms against Gram-positive and Gram-negative bacteria [27]. It is possible that the higher resistance found in Gram-negative bacteria to antimicrobial agents is related to their complex permeability barrier compared to the simpler cell membrane of Gram-positive bacteria. The cell membrane of Gram-negative bacteria has an external lipopolysaccharide barrier that restricts the penetration of most molecules while being permeable to nutrients [28].
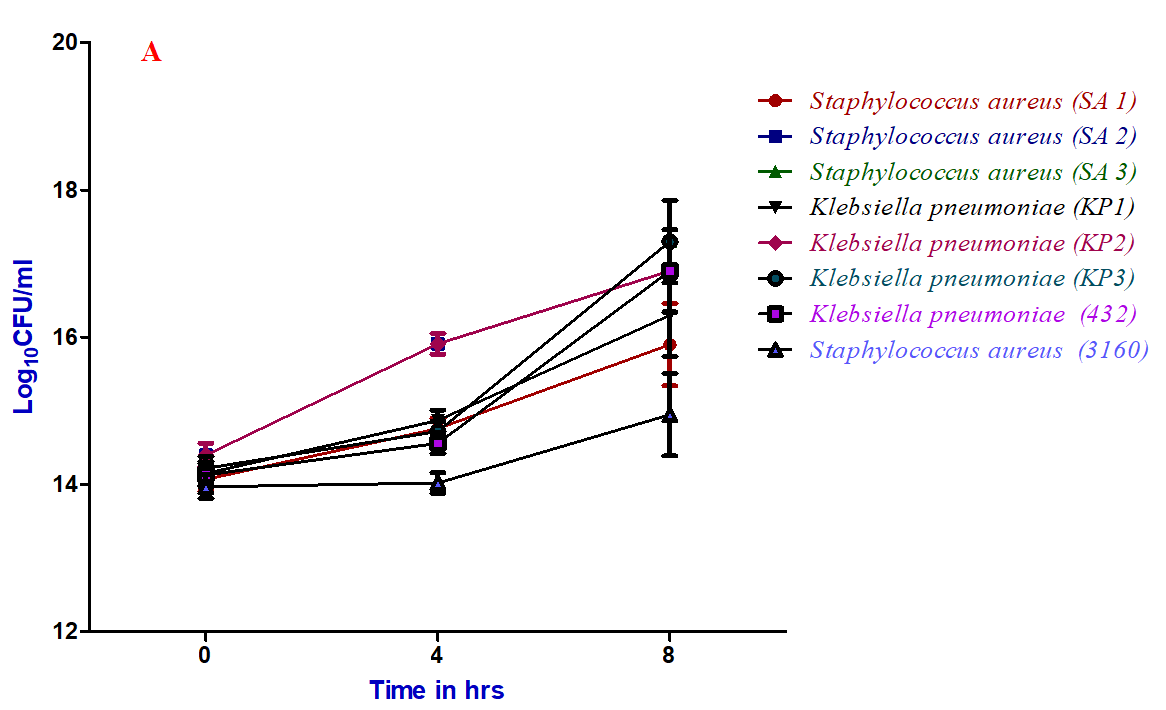
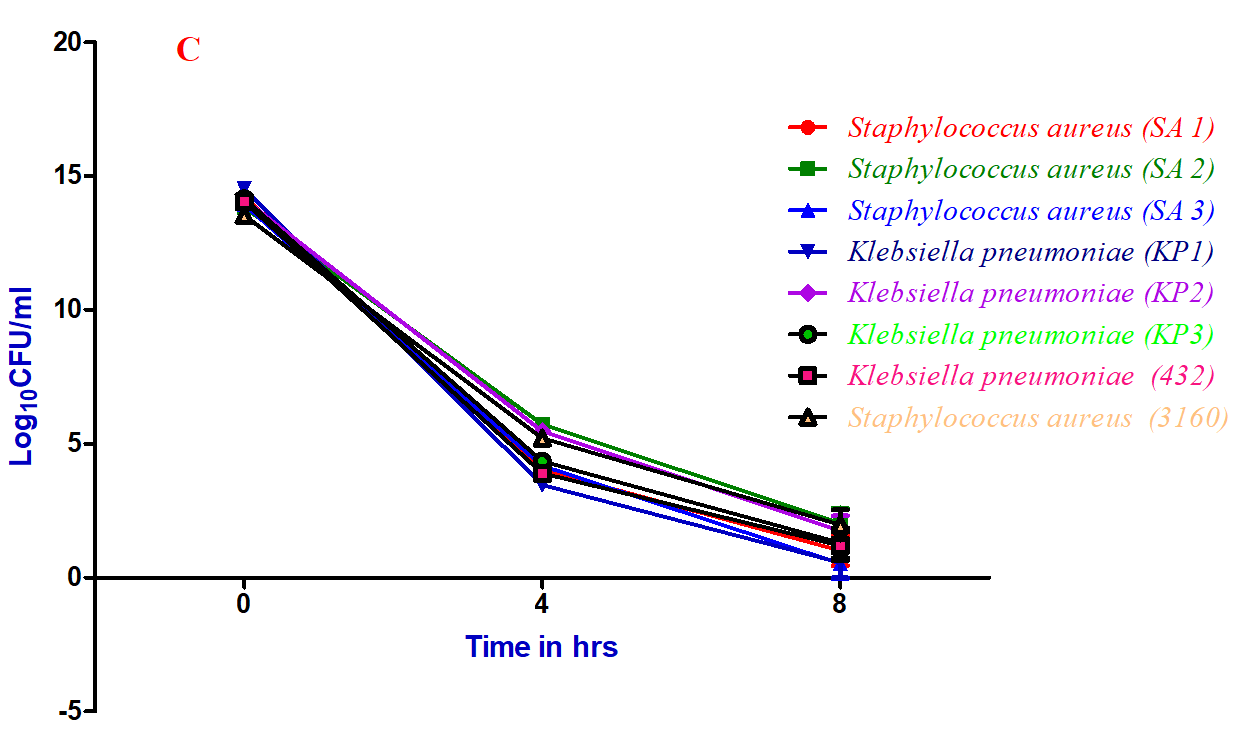
Figure 5: Time-kill kinetic assay of ethyl acetate extract of C. cinerea (C2) culture filtrate against bacterial pathogens: A) 0.5×MIC, B) 1×MIC and C) 2×MIC
Time-kill kinetic assay of ethyl acetate extract of Coprinopsis cinerea (C2) culture filtrate against various strains of Staphylococcus aureus and Klebsiella pneumoniae at different minimum inhibitory concentration (1 and 2 ×MIC) showed a significant reduction in the number of viable cells over 4 and 8hrs of incubation (Figure 5). Whereas, at 0.5 ×MIC, there is no reduction in the number of viable cells even after 8 hrs of incubation. The results clearly showed that half MIC doesn’t have any effect on the reduction of viable cells compared 1 and 2×MIC. However, the course of antibacterial action was observed to be bactericidal and concentration dependent of the specific compound responsible for the same. This is in agreement with the results of Tinrat, who reported the time-kill kinetics of Lentinus edodes, Pleurotus eryngii exhibited bactericidal activity [29].
Conclusion
The results of our study categorically concludes that Coprinopsis cinerea is a potential coprophilous fungus for the production of antibacterial compounds specifically against multi-drug resistant Staphylococcus aureus and Klebsiella pneumoniae when grown under solid state fermentation and ethyl acetate is the suitable and effective solvent for the extraction of the same. Further studies will be carried out to evaluate the chemical nature of the extracts showing antibacterial activity so as to explore possibility of introducing the compound as a drug.
Acknowledgments
The authors would like to thank the Department of Backward classes’ government of Karnataka for providing PHD-OBC fellowship for the first author and also for the Department of Microbiology and Biotechnology, Bangalore University, Bangalore for providing the facility to carry out this research work.
Article Info
Article Type
Research ArticlePublication history
Received: Wed 12, Jun 2019Accepted: Mon 05, Aug 2019
Published: Tue 03, Sep 2019
Copyright
© 2023 J. Savitha. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.CMR.2019.01.01
Author Info
Corresponding Author
J. SavithaDepartment of Microbiology and Biotechnology, Bangalore University, Jnanabharathi campus Bangalore- 560056. Karnataka, India
Figures & Tables
Table 1: Antibiotic susceptibility profile of Gram-positive bacteria (MIC values in µg/ml)
|
Antibiotic |
Staphylococcus aureus (SA 1) |
Staphylococcus aureus (SA 2) |
Staphylococcus aureus (SA 3) |
|||
|
MIC |
INT |
MIC |
INT |
MIC |
INT |
|
|
Benzylpenicillin |
> = 0.5 |
R |
> = 0.5 |
R |
> = 0.5 |
R |
|
Cefoxitin screen |
POS |
+ |
POS |
+ |
POS |
+ |
|
Ciprofloxacin |
>=18 |
R |
>=8 |
R |
>=8 |
R |
|
Clindamycin |
0.25 |
S |
0.25 |
S |
>=4 |
R |
|
Daptomycin |
0.25 |
S |
0.5 |
S |
0.5 |
S |
|
Erythromycin |
1 |
S |
<=0.25 |
S |
>=8 |
R |
|
Gentamicin |
>=16 |
R |
4 |
S |
< = 0.5 |
S |
|
Inducible Clindamycin resistance |
NEG |
- |
NEG |
- |
NEG |
- |
|
Levofloxacin |
4 |
I |
4 |
I |
4 |
1 |
|
Linozolid |
2 |
S |
2 |
S |
2 |
S |
|
Nitrofurantoin |
32 |
S |
< = 16 |
S |
32 |
S |
|
Oxacillin |
> = 4 |
R |
> = 4 |
R |
> = 4 |
R |
|
Rifampicin |
< = 0.03 |
S |
< = 0.03 |
S |
32 |
S |
|
Teicoplanin |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
|
Tetracyclin |
<=1 |
S |
<=1 |
S |
<=1 |
S |
|
Tigecycline |
< = 0.12 |
S |
< = 0.12 |
S |
< = 0.12 |
S |
|
Trimethoprim/Sulfamethoxazole |
>=320 |
R |
80 |
R |
>=320 |
R |
|
Vancomycin |
1 |
S |
< = 0.5 |
S |
1 |
S |
S, Susceptible; I, intermediate; R, resistant (this classification was made according to the interpretative breakpoints suggested by Clinical and Laboratory Standards Institute – CLSI); POS, positive to Cefoxitin screening; NEG, negative to Cefoxitin screening, MIC, Minimum inhibitory concentration, INT, Interpretation
Table 2: Antibiotic susceptibility profile of Gram-negative bacteria (MIC values in µg/ml)
|
Antibiotic |
E. coli (EC 1) |
E. coli (EC 2) |
Klebsiella pneumoniae (KP1) |
Klebsiella pneumoniae (KP2) |
Klebsiella pneumoniae (KP3) |
Pseudomonas aeruginosa |
||||||
|
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
MIC |
INT |
|
|
Amikacin |
< = 2 |
S |
< = 2 |
S |
>=64 |
R |
< = 2 |
S |
>=64 |
R |
>=64 |
R |
|
Amoxicillin/Clavulanic acid |
>=32 |
R |
4 |
S |
>=32 |
R |
>=32 |
R |
>=32 |
R |
na |
|
|
Ampicillin |
>=32 |
R |
>=32 |
R |
>=32 |
R |
>=32 |
R |
>=32 |
R |
na |
|
|
Cefepime |
2 |
S |
16 |
R |
>=64 |
R |
2 |
S |
>=64 |
R |
>=64 |
R |
|
Cefoperazone/Sulbactam |
< = 8 |
S |
16 |
S |
>=64 |
R |
32 |
I |
>=64 |
R |
>=64 |
R |
|
Ceftazidime |
na |
na |
na |
na |
na |
>=64 |
R |
|||||
|
Ceftriaxone |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Cefuroxime |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Cefuroxime Axetil |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
>=64 |
R |
na |
|
|
Ciprofloxacin |
>=4 |
R |
0.5 |
S |
>=4 |
R |
>=4 |
R |
>=4 |
R |
>=4 |
R |
|
Colistin |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
< = 0.5 |
S |
|
Doripenem |
na |
na |
na |
na |
na |
>=8 |
R |
|||||
|
Ertapenem |
< = 0.5 |
S |
< = 0.5 |
S |
>=8 |
R |
< = 0.5 |
S |
>=8 |
R |
na |
|
|
Gentamicin |
< = 1 |
S |
< = 1 |
S |
4 |
S |
>=16 |
R |
< = 1 |
S |
>=16 |
R |
|
Imipenem |
< = 0.25 |
S |
< = 0.25 |
S |
2 |
R |
< = 0.25 |
S |
2 |
R |
>=16 |
R |
|
Levofloxacin |
na |
na |
na |
na |
na |
>=8 |
R |
|||||
|
Meropenem |
< = 0.25 |
S |
< = 0.25 |
S |
>=16 |
R |
< = 0.25 |
S |
>=16 |
R |
>=16 |
R |
|
Minocycline |
na |
na |
na |
na |
na |
>=16 |
R |
|||||
|
Nalidixic acid |
> = 32 |
R |
na |
> = 32 |
R |
na |
na |
na |
||||
|
Nitrofurantoin |
< = 16 |
S |
na |
256 |
R |
na |
na |
na |
||||
|
Piperacillin/Tazobactam |
< = 4 |
S |
8 |
S |
>=128 |
R |
>=128 |
R |
>=128 |
R |
>=128 |
R |
|
Tigecycline |
< = O.5 |
S |
< = O.5 |
S |
> = 8 |
R |
< = O.5 |
S |
4 |
I |
>=8 |
R |
|
Tricarcillin/Clavulanic acid |
na |
na |
na |
na |
na |
>=128 |
R |
|||||
|
Trimethoprim/Sulfamethoxazole |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
>=320 |
R |
interpretative breakpoints suggested by Clinical and Laboratory Standards Institute – CLSI); na, not applicable; POS, positive to Cefoxitin screening; NEG, negative to Cefoxitin screening, MIC, Minimum inhibitory concentration, INT, Interpretation
Table 3: Minimum inhibitory concentration (MIC) and Minimum bactericidal concentration (MBC) of ethyl acetate extract of C. cinerea (C2) against multidrug resistant bacteria (mg/ml)
|
Name of the pathogen |
Solid substrate |
|
|
|
Wheat bran |
|
|
|
MIC |
MBC |
|
Staphylococcus aureus (SA 1) |
3.12 |
6.25 |
|
Staphylococcus aureus (SA 2) |
6.25 |
12.5 |
|
Staphylococcus aureus (SA 3) |
6.25 |
6.25 |
|
Klebsiella pneumoniae (KP1) |
3.12 |
6.25 |
|
Klebsiella pneumoniae (KP2) |
3.12 |
6.25 |
|
Klebsiella pneumoniae (KP3) |
6.25 |
12.5 |
|
Klebsiella pneumoniae (432) |
6.25 |
12.5 |
|
Staphylococcus aureus (3160) |
3.12 |
6.25 |

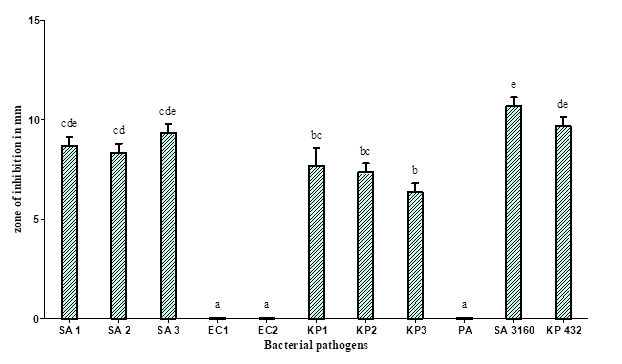


References
- Alves MJ, Ferreira IC, Martins A, Pintado M (2012) Antimicrobial activity of wild mushroom extracts against clinical isolates resistant to different antibiotics. J Appl Microbiol 113: 466-475. [Crossref]
- Shen HS, Shao S, Chen JC, Zhou T (2017) Antimicrobials from mushrooms for assuring food safety. Comprehen Rev Food Sci Food Safety 16: 316-329.
- Bills GF, Gloer JB, An Z (2013) Coprophilous fungi: antibiotic discovery and functions in an underexplored arena of microbial defensive mutualism. Curr Opin Microbiol 16: 549-565. [Crossref]
- Alves MJ, Ferreira IC, Martins A, Pintado M (2012) Antimicrobial activity of wild mushroom extracts against clinical isolates resistant to different antibiotics. J Appl Microbiol 113: 466-475. [Crossref]
- Buller AHR (1931) Further observations on the Coprini together with some investigations on social organization and sex in the hymenomycetes. Res Fung 4.
- Uljé CB, Noordeloos ME (1999) Studies in Coprinus V--Coprinus section Coprinus. Revision of subsection Lanatuli Sing. Mole Phylogen Evolution Fungi 17: 165-199.
- Kües U, Y Liu (2000) Fruiting body production in basidiomycetes. Appl microbiol biotechnol 54: 141-152.
- Mohankumar S, Savitha J (2017) Wheat flour, an inexpensive medium for in vitro cultivation of coprophilous fungus Coprinopsis cinerea. Curr Res Environment Appl Mycol 7: 144-154.
- Yamaç M, Bilgili F (2006) Antimicrobial activities of fruit bodies and/or mycelial cultures of some mushroom isolates. Pharmaceutical biology 44: 660-667.
- Machado CM, Soccol CR, de Oliveira BH, Pandey A (2002) Gibberellic acid production by solid-state fermentation in coffee husk. Appl Biochem Biotechnol 102: 179-191. [Crossref]
- Appiah T, Boakye YD, Agyare C (2017) Antimicrobial activities and time-kill kinetics of extracts of selected ghanaian mushrooms. Evid Based Complement Alternat Med 2017: 4534350. [Crossref]
- Jeyanthi V, Velusamy P (2016) Anti-methicillin resistant Staphylococcus aureus compound isolation from halophilic Bacillus amyloliquefaciens MHB1 and determination of its mode of action using electron microscope and flow cytometry analysis. Indian J Microbiol 56: 148-157. [Crossref]
- Santajit S, Indrawattana N (2016) Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016: 2475067. [Crossref]
- Jonathan SG, Kigigha LT, Ohimain E (2008) Evaluation of the inhibitory potentials of eight higher Nigerian fungi against pathogenic microorganisms. African J Biomed Res 11.
- Eloff JN (1998) Which extractant should be used for the screening and isolation of antimicrobial components from plants? J Ethnopharmacol 60: 1-8. [Crossref]
- Oyetayo VO (2009) Free radical scavenging and antimicrobial properties of extracts of wild mushrooms. Braz J Microbiol 40: 380-386. [Crossref]
- Venturini ME, Rivera CS, Gonzalez C, Blanco D (2008) Antimicrobial activity of extracts of edible wild and cultivated mushrooms against foodborne bacterial strains. J Food prot 71: 1701-1706. [Crossref]
- Barros L, Falcão S, Baptista P, Freire C, Vilas-Boas M et al. (2008) Antioxidant activity of Agaricus sp. mushrooms by chemical, biochemical and electrochemical assays. Food chem 111: 61-66.
- Ozen T, Darcan C, Aktop O, Turkekul I (2011) Screening of antioxidant, antimicrobial activities and chemical contents of edible mushrooms wildly grown in the Black Sea region of Turkey. Comb Chem High Throughput Screen 14: 72-84. [Crossref]
- Solak MH, Kalmis E, Saglam H, Kalyoncu F (2006) Antimicrobial activity of two wild mushrooms Clitocybe alexandri (Gill.) Konr. and Rhizopogon roseolus (Corda) TM Fries collected from Turkey. Phytother Res 20: 1085-1087. [Crossref]
- Dulger B, Ergul CC, Gucin F (2002) Antimicrobial activity of the macrofungus Lepista nuda. Fitoterapia 73: 695-697. [Crossref]
- Karaman M, Jovin E, Malbaša R, Matavuly M, Popović M (2010) Medicinal and edible lignicolous fungi as natural sources of antioxidative and antibacterial agents. Phytother Res 24: 1473-1481. [Crossref]
- Cuadra T, Fernandez FJ, Tomasini A, Barrios‐González J (2008) Influence of pH regulation and nutrient content on cephalosporin C production in solid‐state fermentation by Acremonium chrysogenum C10. Lett Appl Microbiol 46: 216-220. [Crossref]
- El-Naggar MY, El-Assar SA, Abdul-Gawad SM (2009) Solid-state fermentation for the production of meroparamycin by Streptomyces sp. strain MAR01. J Microbiol Biotechnol 19: 468-473. [Crossref]
- Vastrad BM, Neelagund SE (2014) Optimizing the medium conditions for production of tetracycline by solid state fermentation of Streptomyces aureofaciens NCIM 2417 using statistical experimental methods. Biosci Eng 1: 29-44.
- Taofiq O, Heleno SA, Calhelha RC, Alves MJ, Barros L et al. (2016) Development of mushroom-based cosmeceutical formulations with anti-inflammatory, anti-tyrosinase, antioxidant, and antibacterial properties. Molecules 21: E1372. [Crossref]
- Nedelkoska DN, Pančevska NA, Amedi H, Veleska D, Ivanova E, et al. (2013) Screening of antibacterial and antifungal activities of selected Macedonian wild mushrooms. Matica Srpska J Nat Sci 2013: 333-340.
- Oliveira H, Vilas Boas D, Mesnage S, Kluskens LD, Lavigne R et al. (2016) Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front Microbiol 7: 208. [Crossref]
- Tinrat S (2015) Antimicrobial activities and synergistic effects of the combination of some edible mushroom extracts with antibiotics against pathogenic strains. Int J Pharmaceut Sci Rev Res 35: 253-262.