The Potential of U6 and Its Copies in the Regulation of the Human Genome
A B S T R A C T
Non-coding RNAs are conformed by a large repertoire of RNA molecules with unimaginable tridimensional structures and functions. Small nuclear RNAs are an essential part of the spliceosome machinery, which is crucial for proper mRNA maturation. It is important to add that U6, one of the four snRNAs forming the spliceosome has been extensively studied. Full-length U6 (U6-1) loci are widely dispersed throughout the genome (200-900 copies), but a few U6 full-length loci have been identified to date as potentially active genes. The importance of U6 to carry out, together with other snRNAs, the catalytic activity and recognition of annealing target sequences, its evolution in the genome and the fact that the genome has many U6 copies and pseudogenes, its association with retrotransposition, as well as their implication in diseases is discussed in this review.
Keywords
Non-coding RNAs, snRNAs, U6, LINEs, retrotransposition, cancer
Introduction
The human genome sequencing demonstrated that nearly 98-99% of the genome is actively transcribed, but only 1.5% produces a protein product [1-3]. Our genome is constituted by a great variety of functional elements, almost unknown until a few years ago [1, 3]. Furthermore, large fractions of the human transcriptome that do not codify for proteins form an amazing large group of functional non-coding RNAs (ncRNAs) [4]. These versatile molecules have a large repertoire of types and functions that make them key players in the regulation of gene expression and genome stability (Table 1) [4-6].
snRNAs (small nuclear RNAs) are ncRNAs sized ~150 nt in length that have a key function in the spliceosome. In addition, they are crucial for proper mRNA maturation [7]. U6, one of the five snRNAs forming the spliceosome, has been widely studied due to their peculiar promoter region organization, recognized by the Pol III machinery [7, 8]. Besides, contrary to the other snRNAs, which either show gene copies in tandem or are organized in homogeneous repeats, the full-length U6 locus (U6-1, located at 15q23) is widely dispersed throughout the genome [7, 8]. Earlier studies showed ~200 U6 copies dispersed throughout the genome; however, more recent studies estimate ~900 U6 genes copies [9]. Furthermore, it became apparent that the large majority of these copies are U6 pseudogenes, containing nucleotide substitutions and truncations [10]. Only a few U6 copies have been identified to date as potentially active genes; therefore, further studies are necessary to understand the function, if any, of each of these copies and pseudogenes [10]. This poses a challenge, not only due to the number of copies, but also because of the sequence similarities.
Given the large repertoire of types and functions that have been described for ncRNAs, as well as their tissue and development specificity, they have been recently postulated as much more specific markers for pathologies, such as cancer, with respect to mRNA and protein biomarkers [11, 12]. ncRNAs seem to be good molecular tools to improve the classification of types and subtypes of cancer and other pathologies, such as neurodegenerative diseases [11-16]. However, given the high potential of ncRNAs for understanding pathologies, their biogenesis, modes of action, and signaling pathways - both canonical and non-canonical - must be studied in detail.
Table 1: Human ncRNAs types and
functions.
|
Name |
Function |
Length |
|
microRNAs(miRNAs) |
mRNAs
regulation, transcriptional regulators, lncRNAs regulators, Toll receptor
ligands, stabilization of Ago proteins -RISC complex- |
25-100
bp |
|
tRF-derived
RNA fragments |
Regulatory roles
in several biological contexts post-transcriptional
gene regulation. |
40-200
nt |
|
piRNA |
Mostly
involved in the epigenetic and post-transcriptional silencing of transposable
elements. |
26-31
nt |
|
Small
Cajal body-specific RNAs (scaRNA) |
RNA
modification guides |
|
|
Small
nucleolar RNA (snoRNA) |
RNA
modification guides |
60
to 170 nt |
|
Transference
RNA (tRNA) |
Translation-related
RNAs |
76
to 90 nt |
|
YRNA |
Regulatory
RNAs Immunity |
24
to 34nt |
|
Vault
RNA |
Regulatory
RNAs |
88
and 140 nt |
|
Small
nuclear RNA (snRNA) |
Spliceosomal
RNA |
150
nt |
|
RNA
component of mitochondrial RNA processing endoribonuclease (RMRP) |
Catalytic
RNAs |
|
|
Signal
recognition particle RNA (7SL) |
Translation-related
RNAs |
|
|
7SK |
Regulatory
RNAs |
|
|
Ribonuclease
P (RNAseP) |
Catalytic
RNAs |
|
|
Long
non-coding RNAs (lncRNAs) |
Large repertoire
of functions Decoy,
signal, guide, enhancer, scaffold, enzyme regulator, splicing modulation,
small RNA precursor, extracellular secretion, targeting mRNAs, targeting
proteins and miRNAs sponge. |
More
than ~IK200 nt |
|
rRNA |
Translation-related
RNAs |
~1800-5000
nt |
snRNAs
In general, the function of ncRNAs is exerted by base pairing with other nucleic acid molecule. This action can be accompanied by associated proteins also known as ribonucleoprotein complexes [17]. One of the most abundant types of ncRNAs that carry out their function in ribonucleoprotein complexes are snRNAs (sn)RNPs. snRNAs are highly abundant non-coding transcripts, characterized by not being polyadenylated, which carry out their function in the nucleoplasm [17]. These ncRNAs have been categorized in two distinct classes based on their sequences similarity, protein used as cofactors and the polymerase transcribing them: Sm-Class-Pol II and Lsm-class; Pol II-III (Table 2) [7]. Since their discovery as central molecules of the spliceosome machinery, further works have been mainly focused on the study of snRNAs biology and their mode of action. These studies showed a high complexity in snRNAs assembly, trafficking, and mechanisms of action [7, 17].
Table 2: snRNAs classes: Sm-Class-Pol II and Lsm-class; Pol
II-III.
|
snRNA
class |
snRNA |
Maturation |
Processed
by |
Structure |
|
snRNA Sm |
U1, U2, U4, U4atac, U5,
U7, U11 and U12 |
Subnuclear
structures. They need to leave the nucleus and return to it. |
Transcribed by
Pol II. It requires the union of general transcript factors, such as TFIIA,
TFIIB, TFIIE and TFIIF. |
Constituted by a
5′-trimethylguanosine cap, a 3′stem–loop and a hetero-heptameric ring
structure -Sm site- proteins binding site. |
|
snRNA Lsm |
U6 and U6atac |
Only in subnuclear
structures-they never leave the nucleus- |
Transcribed by
Pol III |
Constituted by a
monomethylphosphate cap and a 3′stem- loop, terminating in a stretch of
uridines - proteins binding site. |
I The Spliceosome
The spliceosome is a ribonucleoprotein complex essential for the correct mRNA maturation, constituted by five different subunits of uridine-rich small snRNAs (U1, U2, U4, U5 and U6), by the canonical splicing machinery or (U11, U12, U6atac) non-canonical splicing machinery, by more than 150 proteins, interacting with snRNAs denominated snRNPs, and by many others accessory proteins [7, 8]. All these make up a specialized macro-ribonucleoprotein complex mediating intronic cleavage. Both the spliceosome biogenesis and function are regulated by specific chemical modifications regulating base-pairing interactions among snRNAs [7, 8, 18, 19].
II RNU6
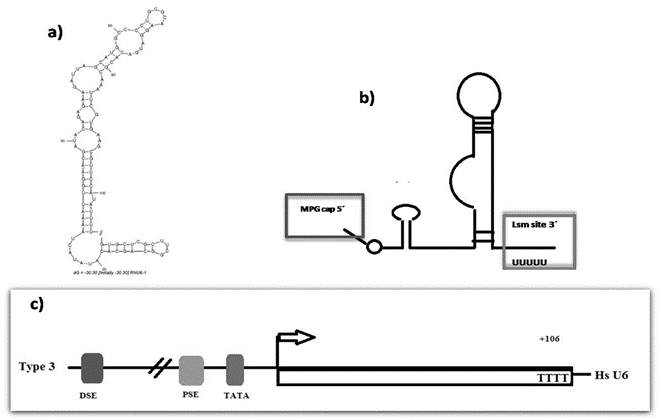
RNU6 or U6 (U6-1) is a uridine-rich small non-coding RNA sized ~ 61 nt in length. Interestingly, it is an essential ncRNA for nuclear intron splicing (Figure 1), having structural and functional similarities to that of the domain V of autocatalytic group II introns [7, 8, 20, 21]. The similarity with the V domain is observed in the AGC triad, forming the U2-U6 fold, which is present and conserved at both genomic elements, favouring the same tertiary folding interactions. Group II introns are present in bacteria, archaebacteria, mitochondria, and chloroplasts, but are notably excluded from nuclear genomes [20, 21]. The degree of conservation of the secondary and tertiary structures between group II introns and the U2-U6 fold at the active site level strongly suggest that these introns and the spliceosome share a common evolutionary origin [22]. In addition, GU oscillations at the beginning of the U2-U6 fold and during the formation of the catalytic center of group II introns have been observed [21, 22]. U6 is the most conserved spliceosomal snRNA with more than a billion years of eukaryotic evolution [23].
Figure 1: Putative secondary structure of human U6 and the structure of the promoter gene. a) Secondary structure of human U6 accordingly to The RNA Fold Web Server [59]. b) Secondary structure of human U6 accordingly to Didychuk et al., showing the 5′ γ-monomethyl cap and the 3′ U-tail present in Lsm class. c) Essential upstream elements of U6 promoter including TATA box 30 base pairs upstream of the transcription start site, a proximal sequence element (PSE) 50 base pairs upstream, and a distal sequence element (DSE) 250 base pairs upstream [59].
RNU6 Interactome
RNU6 can interact with different molecules through the splicing cycle - three snRNAs; the pre-mRNA substrate; and more than 25 protein partners. Therefore, RNU6 is a highly dynamic molecule [7, 8, 19, 21, 23]. These interactions occur throughout extensive structural U6 rearrangements, including unwinding and reformation of stable internal secondary structure, which are attributed to U6 post-transcriptional modifications during its biogenesis [8]. However, the precise effect of each modification has not been fully elucidated [8, 24]. Interestingly, U6 is located in the core of the catalytic site, and it binds the magnesium ion needed for the spliceosomal machinery, regulating together with U2 and U5 the position of the substrate, which is fundamental for the reaction [7, 8, 19, 21, 23].
Genomic Organization
Regarding the genomic structure and organization, U6 differs from other snRNAs not only in the type of promoter controlling its transcription, but also in the number of copies at the genome [25, 26]. For example, U1 and U2 are highly conserved genes, having 10-30 copies of true genes coding for snRNAs. Meanwhile, human U6 has 200-900 copies coding for pseudogenes with many nucleotide substitutions and full-length truncations scattered throughout the genome and with unknown functions [9, 10].
U6 Promoter
Type III promoters – recognized by Pol III - were identified for the first time in the mammalian U6 snRNA gene and in the human 7SK gene [27-29]. We have gained more knowledge about this kind of promoter since approximately 30 years ago. Few genes of our genome are recognized by Pol III, however, most of them are RNAs: catalytic, structural, and/or with unknown function, which are involved mainly in cell growth and cell cycle regulation [29, 30].
Based on the promoter region, specifically in the 5’ terminal region, genes type 3 are divided in three distinct categories: i) promoter-gene-internal, ii) generally TATA-less, as those found in the VAI and tRNA genes and; iii) gene-external, containing a TATA box, as exemplified by the U6 snRNA gene (Figure 1) [30]. Furthermore, the specificity of Pol III can change and alternate with Pol II through mechanisms, such as TATA box elimination or addition [9-26, 31]. It is believed that the recruitment of certain transcription factors used by Pol II and that are also necessary for PoI III influences chromatin remodeling. This, in turn allows the interaction of both polymerases [30]. Additionally, although the U6 gene is highly conserved among organisms, its Pol III promoter structure is divergent. Studies performed in the human U6 gene demonstrated the interaction between Pol II and Pol III and that this interaction at a site ~300 bp upstream is essential for its transcription [27-38]. Remarkably, the transcription of U6atac is also dependent on both Pol II and Pol III [31, 36]. Despite these, further studies are necessary to evaluate the transcriptional activity of U6 and its involvement in different tissues and development stages and pathologies, given that specific tissue differences in the expression of U6 have been reported [23, 25].
Mechanisms of U6 Retrotransposition
Although the genome has ‘allowed’ and maintained many repeated sequences and copy number expansions, it is difficult to know the exact function they exert. However, the presence of these sequences along the human evolution has shaped the genome and new functional elements have been generated in most cases by a unique mechanism called retrotransposition [39]. Retrotransposition is the greatest remodeling force of the human genome, and it is currently represented by the Long Interspersed Nuclear Element 1 (LINE 1) [40-43]. These are 6-7kb elements that have all the machinery to be copied and mobilized into the genome. Generally, the 5’UTR has two promoters; the sense promoter directs the transcription of ORF1 and ORF2; and ASP directs the transcription of ORF0. The 3’UTR harbors both Alu and SVS elements, and the LINE 1 machinery to regulate its mobilization. The intrinsic nature of LINE 1 is to be copied and inserted itself into the genome, increasing genomic instability. In response to this, the cell has ‘engineered’ many mechanisms that control the levels of LINE1 retrotransposition; alterations in these regulatory systems can increase the mobility of LINE-1 and promote the formation of chimeric genes; however, in the case of U6, the conundrum is why a large spread of U6 has been ‘allowed’ in the human genome [43].
In the 80s, the first studies of snRNA pseudogenes formation showed 4 different classes among which, three classes could be formed by using RNA as intermediate [43-47]. Recent studies have redefined the snRNAs pseudogene formation classification based on their structure organization (Table 3) Doucet et al. 2015 described in detail some processes by which LINE-1 elements can recruit RNA molecules, especially the U6 sequence, promoting diversification and propagation of these sequences in the genome, not only in humans, but also in different species; however, the largest number of inserted and scattered copies are found in the human genome [42]. Interestingly, computational analysis demonstrated that the three LINE clades (L1, L2, and RTE) were able to form U6 pseudogenes, even when L1 is actually the only active element in the genome, indicating this phenomenon has occurred throughout evolution. Remarkably, this has favoured the diversification of these sequences [42].
Table 3:
Pseudogenes structure organization.
|
Group |
Characteristics |
Mechanism of
amplification |
|
Group I |
Duplications of snRNA genes and their flanking
sequence. |
|
|
Group II |
Processed snRNA pseudogenes that generally end
with an A-rich tail and are flanked by TSD. |
Reverse transcription initiates directly on the
snRNA by a template choice. |
|
Group III |
Processed pseudogenes that are heavily
30-truncated and flanked by TSD. |
Reverse transcription initiates directly on the
snRNA by a template choice. |
|
Group IV |
snRNA pseudogenes that form chimeras with a
non-LTR retrotransposon. |
Template switching. |
Moldovan et al. reported the U6/L1 chimera can be generated more than once every fifteen retrotransposition events in HeLa cells, which according to the author it may suggest that the formation of U6/L1 chimera is a common event in the genome [47]. The USB1 (U6 snRNA phosphodiesterase) gene acts as exoribonuclease (RNase) responsible for trimming the (polyU) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA 3’ end terminated with a 2′,3′-cyclic phosphate [48]. This terminal region is important because the authors demonstrated that the enzyme RTCB (RNA 2',3'-Cyclic Phosphate and 5'-OH Ligase) can facilitate the replacement of the 5’ region of U6/L1 chimeras, mediated by the elimination of the 2′,3′-cyclic phosphate.
Interestingly, U6/L1 chimeric RNAs can arise independently of L1 retrotransposition and are formed through the 2′,3′-cyclic phosphate ligation on the 3′ end of the U6 snRNA and 5′-OH on L1 RNA [48]. Chimeric U6/L1 RNAs are an important component of the transcriptome in multiple human cell lines [48]. As mentioned by Doucet et al., the fact that U6 has many copies and variants in the human genome has posed a challenge for the study of the possible role of each of these copies and variants. It is highly likely that these copies and variants have been omitted in many studies due to the lack of annotations in databases [48]. Many of these sequences have poor conservation among organisms and this is an important aspect to further understand their biological function [49]. Nevertheless, it has been observed in lncRNAs, which represent the less conserved and more recent sequences in the human genome with respect to other organisms, that although they do not keep their sequence identity, they maintain their function and are specifically transcribed in a variety of organisms [35, 49]. Importantly, recent findings regarding ‘pseudogenes’ have demonstrated the high regulatory capacity these sequences can exert in the genome [50]. So far, the genome has shown us that most of its transcripts are used to regulate gene expression in a very fine way. For this reason, the fact that the genome has been overly concerned with maintaining U6 in mammals giving variability is challenging.
It has been reported that at least four U6 human genes are transcriptionally active and the expression of a U6 variant with nine substitutions and one deletion can be expressed under the control of an internal promoter [38, 39]. U6atac, a U6 paralogue gene, proves that even when these sequences have a very low identity than other copies of U6, they can play a critical role in the cell. U6atac is a cofactor in the splicing of AT ± AC type introns and it is expressed at low levels in human cells [51, 52]. Based on all these, it can be postulated that many of the scattered copies of U6 at the genome could also be having a biological function.
U6 Involvement in Diseases
U6 has been associated with some diseases, for example, mutations in USB1 (U6 SnRNA Biogenesis Phosphodiesterase 1), which is an exoribonuclease essential for the modification of spliceosomal small nuclear RNA (snRNA) U6 by trimming its oligouridine tail and introducing a cyclic phosphate group, has been associated with accelerated U6 decay and pre-mRNA splicing defects [53, 54]. It is likely that this deficiency induces U6 3′ end misprocessing. Mutations in this gene are associated with the genodermatosis Clericuzio-type poikiloderma with neutropenia (PN) [53, 54]. In a seemingly contradictory way, the USB1 knockdown in HeLa cells showed no alteration in the expression levels of U6 and an apparently correct pre-mRNA splicing; however, U6 snRNA molecules became extended with a more heterogeneous length compared with controls. USB1 loss also modestly decreased U6 snRNA stability [53, 55]. Accordingly with Moldovan et al., the 3′ region is important because, as mentioned above, the enzyme RTCB can facilitate the replacement of this region of U6/L1 chimeras, mediated by the elimination of the 2′,3′-cyclic phosphate. The real implication of the modification exerted by USB1 in this terminal domain remains to be elucidated.
Cancer
It has been reported in murine models of cancer an increased Pol III activity, which was later corroborated with transformed human cells, showing an increased transcript expression controlled by Pol III [56]. This seems to be strongly related to cellular transformation [56]. The latter is not surprising, since Pol III controls gene expression of transcripts involved mainly in cell growth and cell cycle regulation [27-30]. Importantly, Cabarcas et al. 2010, showed that U6 transcription inhibition was mediated by PTEN (Phosphatase and Tensin Homolog) through the lipid-binding C2 domain of PTEN, and its interaction with BRF2 (BRF2 RNA Polymerase III Transcription Initiation Factor Subunit) [57]. Meanwhile, Puigdelloses et al., showed the U6 overexpression in the blood serum of adult patients with GBM (glioblastoma multiforme) and postulated this snRNA as potential biomarker to differentiate GBM from brain lesions that cannot be detected by imaging [58]. Due to cellular and cell growth regulation processes in which RNU6 is involved, an increase in its expression seems to be essential for tumor growth [27-30, 56, 57]. Therefore, it is necessary to conduct further studies regarding the functions performed by U6 to regulate cancer, as well as the functions performed by each U6 copy and if there is a tissue-specific expression related to some pathology.
Conclusion
U6 is undoubtedly an snRNA with essential functions in the human genome, since during evolution it has considerably increased its number of copies and variants in the human genome. Although the study of U6 represents a challenge for the scientific community, we firmly believe that it will elucidate many important functions of this RNA in health and in disease.
Acknowledgment
The financial resources to carry out this work were provided by the authors.
Conflicts of Interest
None.
Competing Interests
None.
Article Info
Article Type
Review ArticlePublication history
Received: Thu 19, Aug 2021Accepted: Thu 02, Sep 2021
Published: Thu 16, Sep 2021
Copyright
© 2023 Ruth Ruiz Esparza-Garrido. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.COR.2021.09.05
Author Info
Miguel Angel Velazquez-Flores Ruth Ruiz Esparza-Garrido
Corresponding Author
Ruth Ruiz Esparza-GarridoLaboratorio de RNAs no codificantes de la Unidad de Investigacion Medica en Genetica Humana, Hospital de Pediatria, CMNSXXI, Instituto Mexicano del Seguro Social (IMSS), Mexico City, Mexico
Figures & Tables
Table 1: Human ncRNAs types and
functions.
|
Name |
Function |
Length |
|
microRNAs(miRNAs) |
mRNAs
regulation, transcriptional regulators, lncRNAs regulators, Toll receptor
ligands, stabilization of Ago proteins -RISC complex- |
25-100
bp |
|
tRF-derived
RNA fragments |
Regulatory roles
in several biological contexts post-transcriptional
gene regulation. |
40-200
nt |
|
piRNA |
Mostly
involved in the epigenetic and post-transcriptional silencing of transposable
elements. |
26-31
nt |
|
Small
Cajal body-specific RNAs (scaRNA) |
RNA
modification guides |
|
|
Small
nucleolar RNA (snoRNA) |
RNA
modification guides |
60
to 170 nt |
|
Transference
RNA (tRNA) |
Translation-related
RNAs |
76
to 90 nt |
|
YRNA |
Regulatory
RNAs Immunity |
24
to 34nt |
|
Vault
RNA |
Regulatory
RNAs |
88
and 140 nt |
|
Small
nuclear RNA (snRNA) |
Spliceosomal
RNA |
150
nt |
|
RNA
component of mitochondrial RNA processing endoribonuclease (RMRP) |
Catalytic
RNAs |
|
|
Signal
recognition particle RNA (7SL) |
Translation-related
RNAs |
|
|
7SK |
Regulatory
RNAs |
|
|
Ribonuclease
P (RNAseP) |
Catalytic
RNAs |
|
|
Long
non-coding RNAs (lncRNAs) |
Large repertoire
of functions Decoy,
signal, guide, enhancer, scaffold, enzyme regulator, splicing modulation,
small RNA precursor, extracellular secretion, targeting mRNAs, targeting
proteins and miRNAs sponge. |
More
than ~IK200 nt |
|
rRNA |
Translation-related
RNAs |
~1800-5000
nt |
Table 2: snRNAs classes: Sm-Class-Pol II and Lsm-class; Pol
II-III.
|
snRNA
class |
snRNA |
Maturation |
Processed
by |
Structure |
|
snRNA Sm |
U1, U2, U4, U4atac, U5,
U7, U11 and U12 |
Subnuclear
structures. They need to leave the nucleus and return to it. |
Transcribed by
Pol II. It requires the union of general transcript factors, such as TFIIA,
TFIIB, TFIIE and TFIIF. |
Constituted by a
5′-trimethylguanosine cap, a 3′stem–loop and a hetero-heptameric ring
structure -Sm site- proteins binding site. |
|
snRNA Lsm |
U6 and U6atac |
Only in subnuclear
structures-they never leave the nucleus- |
Transcribed by
Pol III |
Constituted by a
monomethylphosphate cap and a 3′stem- loop, terminating in a stretch of
uridines - proteins binding site. |
Table 3:
Pseudogenes structure organization.
|
Group |
Characteristics |
Mechanism of
amplification |
|
Group I |
Duplications of snRNA genes and their flanking
sequence. |
|
|
Group II |
Processed snRNA pseudogenes that generally end
with an A-rich tail and are flanked by TSD. |
Reverse transcription initiates directly on the
snRNA by a template choice. |
|
Group III |
Processed pseudogenes that are heavily
30-truncated and flanked by TSD. |
Reverse transcription initiates directly on the
snRNA by a template choice. |
|
Group IV |
snRNA pseudogenes that form chimeras with a
non-LTR retrotransposon. |
Template switching. |
References
1. Lander ES, Linton
LM, Birren B, Nusbaum C, Zody MC et al. (2001) Initial sequencing and
analysis of the human genome. Nature 409: 860-921. [Crossref]
2. Venter JC, Adams
MD, Myers EW, Li PW, Mural RJ et al. (2001) The sequence of the human genome. Science
291: 1304-1351. [Crossref]
3. Goffeau A, Barrell
BG, Bussey H, Davis RW, Dujon B et al. (1996) Life with 6000 genes. Science
274: 546, 563-567. [Crossref]
4. Jarroux J, Morillon
A, Pinskaya M (2017) History, Discovery, and Classification of lncRNAs. Adv
Exp Med Biol 1008: 1-46. [Crossref]
5. ENCODE Project
Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R et
al. (2007) Identification and analysis of functional elements in 1% of the
human genome by the ENCODE pilot project. Nature 447: 799-816. [Crossref]
6. ENCODE Project
Consortium (2012) An integrated encyclopedia of DNA elements in the human
genome. Nature 489: 57-74. [Crossref]
7. Matera AG, Terns
RM, Terns MP (2007) Non-coding RNAs: lessons from the small nuclear and small
nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209-220. [Crossref]
8. Didychuk AL,
Butcher SE, Brow DA (2018) The life of U6 small nuclear RNA, from cradle to
grave. RNA 24: 437-460. [Crossref]
9. Mattaj IW, Dathan
NA, Parry HD, Carbon P, Krol A et al. (1988) Changing the RNA polymerase
specificity of U snRNA gene promoters. Cell 55: 435-442. [Crossref]
10. Domitrovich AM,
Kunkel GR (2003) Multiple, dispersed human U6 small nuclear RNA genes with
varied transcriptional efficiencies. Nucleic Acids Res 31: 2344-2352. [Crossref]
11. Deng X, Berletch
JB, Nguyen Di K, Disteche CM (2014) X chromosome regulation: diverse patterns
in development, tissues and disease. Nat Rev Genet 15: 367-378. [Crossref]
12. Grillone K, Riillo C, Scionti F, Rocca R, Tradigo G et al. (2020) Non-coding
RNAs in cancer: platforms and strategies for investigating the genomic “dark
matter”. J Exp Clin Cancer Res 39: 117. [Crossref]
13. Esteller M (2011)
Non-coding RNAs in human disease. Nat Rev Genet 12: 861-874. [Crossref]
14. Arun G, Diermeier
SD, Spector DL (2018) Therapeutic Targeting of Long-Noncoding RNAs in Cancer. Trends
Mol Med 24: 257-277. [Crossref]
15. Gupta SC, Tripathi
YN (2017) Potential of long non-coding RNAs in cancer patients: from biomarkers
to therapeutic targets. Int J Cancer 140: 1955-1967. [Crossref]
16. Anastasiadou E,
Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer
18: 5-18. [Crossref]
17. Huttenhofer A,
Schattner P (2006) The principles of guiding by RNA: chimeric RNA-protein
enzymes. Nat Rev Genet 7: 475-782. [Crossref]
18. Wahl MC, Will CL,
Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell
136: 701-718. [Crossref]
19. Will CL, Lührmann R
(2011) Spliceosome structure and function. Cold Spring Harb Perspect Biol
3: a003707. [Crossref]
20. Fica SM, Tuttle N, Novak T, Sheng Li N, Lu J et al. (2013) RNA
catalyses nuclear pre-mRNA splicing. Nature 503: 229-234. [Crossref]
21. Keating KS, Toor N,
Perlman PS, Pyle AM (2010) A structural analysis of the group II intron active
site and implications for the spliceosome. RNA 16: 1-9. [Crossref]
22. Newby MI, Greenbaum
NL (2001) A conserved pseudouridine modification in eukaryotic U2 snRNA induces
a change in branch-site architecture. RNA 7: 833-845. [Crossref]
23. Tarn WY, Steitz JA
(1996) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare
AT±AC introns. Science 273: 1824-1432. [Crossref]
24. Fu XD (2014)
Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev 1:
190-204. [Crossref]
25. Tichelaar JW,
Wieben ED, Reddy R, Vrabel A, Camacho P et al. (1998) In vivo expression of a
variant human U6 RNA from a unique,internal promoter. Biochemistry 37:
12943-12951. [Crossref]
26. Matera AG, Wang Z
(2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15:
108-121. [Crossref]
27. Krol A, Carbon P,
Ebel JP, Appel B (1987) XenopustropicalisU6
snRNA genes transcribed by Pol III contain the upstream promoter elements used
by Pol II dependent UsnRNAgenes. Nucleic
Acids Res 15: 2463-2478. [Crossref]
28. Kunkel GR, Pederson
T (1988) Upstream elements required for efficient transcription of a human U6
RNA gene resemble those of U1 and U2 genes even though a different polymerase
is used. Genes Dev 2: 196-204. [Crossref]
29. Murphy S, Tripodi
M, Melli M (1986) A sequence upstream from the coding region is required for
the transcription of the 7SK RNA genes. Nucleic
Acids Res 14: 9243-9260. [Crossref]
30. Schramm L,
Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes
Dev 16: 2593-2620. [Crossref]
31. Hernandez N, Lucito
R (1988) Elements required for transcription initiation of the human U2 snRNA
gene coincide with elements required for snRNA 3' end formation. EMBO J 7: 3125-3134. [Crossref]
32. Lobo SM, Hernandez
N (1989) A 7 bp mutation converts a human RNA polymerase II snRNA promoter into
an RNA polymerase III promoter. Cell 58: 55-67. [Crossref]
33. Das G, Henning D,
Wright D, Reddy R (1988) Upstream regulatory elements are necessary and
sufficient for transcription of a U6 gene by RNA polymerase III. EMBO J 7: 503-512. [Crossref]
34. Listerman I, Bledau
AS, Grishina I, Neugebauer KM (2007) Extragenic Accumulation of RNA Polymerase
II Enhances Transcription by RNA Polymerase III. PLoS Genet 3: e212. [Crossref]
35. Lunyak VV, Atallah
M (2011) Genomic relationship between SINE retrotransposons, Pol III-Pol II
transcription, and chromatin organization: the journey from junk to jewel. Biochem
Cell Biol 89: 495-504. [Crossref]
36. Raha D, Wang Z,
Moqtaderi Z, Wu L, Zhong G et al. (2010) Close association of RNA polymerase II
and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A
107: 3639-3644. [Crossref]
37. Hett A, West S
(2014) Inhibition of U4 snRNA in human cells causes the stable retention of
polyadenylated pre-mRNA in the nucleus. PLoS One 9: e96174. [Crossref]
38. Mattaj IW, Hamm J
(1989) Regulated splicing in early development and stage-specific U snRNPs. Development
105: 183-189. [Crossref]
39. Hoeppner MP,
Denisenko E, Gardner PP, Schmeier S, Poole AM (2018) An Evaluation of Function
of Multicopy Noncoding RNAs in Mammals Using ENCODE/FANTOM Data and Comparative
Genomics. Mol Biol Evol 35: 1451-1462. [Crossref]
40. Hayashi K (1981)
Organization of sequences related to U6 RNA in the human genome. Nucleic
Acids Res 9: 3379-3388. [Crossref]
41. Tichelaar JW,
Knerer B, Vrabel A, Wieben ED (1994) Transcription of a variant human U6 small
nuclear RNA gene is controlled by a novel, internal RNA polymerase III
promoter. Mol Cell Biol 14: 5450-5457. [Crossref]
42. Doucet AJ, Droc G,
Siol O, Audoux J, Gilbert N (2015) U6 snRNA Pseudogenes: Markers of
Retrotransposition Dynamics in Mammals. Mol Biol Evol 32: 1815-1832. [Crossref]
43. Cervantes Ayalc A,
Ruiz Esparza Garrido R, Velázquez Flores MÁ (2020) Long Interspersed Nuclear
Elements 1 (LINE1): The chimeric transcript L1-MET and its involvement in
cancer. Cancer Genet 241: 1-11. [Crossref]
44. Van Arsdell SW,
Denison RA, Bernstein LB, Weiner AM, Manser T et al. (1981) Direct repeats
flank three small nuclear RNA pseudogenes in the human genome. Cell 26:
11-17. [Crossref]
45. Van Arsdell SW,
Weiner AM (1984) Pseudogenes for human U2 small nuclear RNA do not have a fixed
site of 3' truncation. Nucleic Acids Res 12: 1463-1471. [Crossref]
46. Denison RA, Weiner
AM (1982) Human U1 RNA pseudogenes may be generated by both DNA- and
RNA-mediated mechanisms. Mol Cell Biol 2: 815-828. [Crossref]
47. Moldovan JB, Wang
Y, Shuman S, Mills RE, Moran JV et al. (2019) RNA ligation precedes the
retrotransposition of U6/LINE-1 chimeric RNA. Proc Natl Acad Sci U S A
116: 20612-20622. [Crossref]
48. Lund E, Dahlberg JE
(1992) Cyclic 2',3'-phosphates and nontemplated nucleotides at the 3' end of
spliceosomal U6 small nuclear RNA's. Science 255: 327-330. [Crossref]
49. Brow DA, Guthrie C
(1988) Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature
334: 213-218. [Crossref]
50. Poliseno L,
Marranci A, Pandolfi PP (2015) Pseudogenes in Human Cancer. Front Med
(Lausanne) 2: 68. [Crossref]
51. Shukla GC, Cole AJ,
Dietrich RC, Padgett RA (2002) Domains of human U4atac snRNA required for
U12-dependent splicing in vivo. Nucleic Acids Res 30: 4650-4657. [Crossref]
52. Tarn WY, Steitz JA
(1977) Pre-mRNA splicing: the discovery of anew spliceosome doubles the
challenge. Trends Biochem Sci 22: 132-137. [Crossref]
53. Mroczek S, Krwawicz
J, Kutner J, Lazniewski M, Kuciński I et al. (2012) C16orf57, a gene mutated in
poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible
for the U6 snRNA 3' end modification. Genes Dev 26: 1911-1925. [Crossref]
54. Arnold AW, Itin PH,
Pigors M, Kohlhase J, Bruckner Tuderman L et al. (2010) Poikiloderma with
neutropenia: a novel C16orf57 mutation and clinical diagnostic criteria. Br J Dermatol 163: 866-869. [Crossref]
55. Mroczek S,
Dziembowski A (2013) U6 RNA biogenesis and disease association. Wiley
Interdiscip Rev RNA 4: 581-592. [Crossref]
56. Marshall L,
Goodfellow SJ, White RJ (2007) Diminished activity of RNA polymerase III
selectively disrupts tissues with the most actively dividing cells. PLoS
Biol 5: e286. [Crossref]
57. Cabarcas S, Watabe
K, Schramm L (2010) Inhibition of U6 snRNA Transcription by PTEN. Online J
Biol Sci 10: 114-125. [Crossref]
58. Puigdelloses M, González Huárriz M, García Moure M, Martínez Vélez N, Esparragosa Vázquez I et al. (2020) RNU6-1 in circulating exosomes differentiates GBM from non-neoplastic brain lesions and PCNSL but not from brain metastases. Neurooncol Adv 2: vdaa010. [Crossref]
59. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406-3415. [Crossref]
