Survival Analysis and Prognostic Predictor Study of Colorectal Cancer Patients with Single-Site Metastasis
A B S T R A C T
Metastatic colorectal cancer (mCRC) patients have various metastasis patterns, which reflect diverse biological characteristics of different patient subgroups. We analyse the prognosis of mCRC patients according to the metastatic site and clarify the relationship between tumor or patient characteristics and the metastatic sites. The whole sequencing and clinical data of 2329 CRC patients were obtained from TCGA and a database of the MSKCC. Kruskal Wallis Tests were used to analyse measurement data. Survival was illustrated by Kaplan-Meier curves, with P value determined by Log-rank Test. Hazard’s ratio was determined through the univariate and multivariate COX proportional hazards regression model. The mortality rate of CRC patients with liver-only metastasis (mCRC-liver) did not increase versus non-metastatic patients. The survival rate of patients with non-regional lymph node-only metastasis (mCRC-NRLN) was lower versus mCRC-liver. Mutations of KRAS and TCF7L2 genes were associated with mortality of mCRC-liver. APC mutation was associated with reduced mortality in mCRC-lung and mCRC-NRLN. BRAF mutation was associated with increased mortality of mCRC-peritoneum. In a multivariate COX analysis, gender affected the survival rate of mCRC-liver. Age and the number of gene mutations affected the survival rate of mCRC-lung and mCRC-NRLN respectively. Receiving chemotherapy is an unfavourable factor for prognosis of mCRC-liver, but the length of chemotherapy treatment is an advantageous prognosis factor. This study depicts the long-term survival features of a group of mCRC patients. These findings promoted our understanding of the prognosis characteristics of CRC and have positive guiding significance for clinical management of CRC patients.
Keywords
Colorectal cancer, metastatic colorectal cancer, prognostic predictor, metastasis, survival rate
Graphical Abstract
Schematic diagram summarizing factors closely related to the overall survival of CRC patients with different single site metastasis.
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide [1]. A variety of CRC patients undergo significant clinical events, including post-diagnostic recurrences or metastases. In spite of recent progress in early detection, systematic treatment and localized therapy, CRC is still one of the major causes of cancer-related deaths and morbidity in the world [2-5]. Approximately 694,000 deaths from CRC occurred worldwide in 2012 [6]. Untreated metastatic colorectal cancer (mCRC) patients have poor prognosis with an average survival time of 5-6 months [7]. Therefore, the assessment of the features of potential disease outcomes and the identification of their predictors are crucial for efficacious patient surveillance and for the treatment and control of this disease in both the short and long term.
A large number of CRC-related deaths are caused by metastatic disease. About 20% of patients had metastatic disease at the time of diagnosis [8]. Although liver, lungs and peritoneum are the most prominent sites of CRC metastasis, there are many other sites of metastasis, e.g. bones, brain and distant lymph nodes [9-12]. Autopsy studies investigating metastasis patterns revealed that distinctive primary cancers metastasize to different sites with different frequencies, and different CRC patients have different spread patterns [12]. Metastasis number and tumor volume are important indicators to predict prognosis [13]. As to whether the anatomical location of the metastasis and the origin of the primary lesion have any influence on the prognosis, there is no final conclusion at present [13, 14]. Elias et al. reported that there was no correlation between metastasis site and prognosis, while Carpizo et al. observed a significant correlation [13, 15]. Nevertheless, both studies are limited by the small sample size. The survival rate of CRC patients with lung-only metastases seems to be at least equivalent to that of patients with liver only metastases, while the prognosis of patients with bone, peritoneal and brain metastases is much worse [16-18]. Two additional studies focus on the survival pattern of mCRC patients, but one covers various patients before modern chemotherapy and the other is much smaller in scale [19, 20]. It is possible that distinct genotypes of mCRC may have different patterns of progression, malignancy and prognosis. We performed the current research to investigate the survival of mCRC patients with different metastatic sites at the time of diagnosis.
Furthermore, it has increasingly been accepted that the biological behaviour and prognosis of CRC may differ depending on the location of the primary lesion and the status of specific mutations, (KRAS, BRAF, and MMR), although the data are not consistent [19, 21]. Several retrospective analysis shows that these somatic and germ line mutations might affect the metastasis pattern, however, most of these are limited, uncontrolled single-agency studies with various correlations [22, 23]. The second aim of the study explored the relationship between molecular markers, primary sites and metastatic sites.
Although CRC is a common disease worldwide, its long-term prognosis characteristics and predictive factors are still unclear. In the present study, we analysed 23 years of data obtained from a prospective patient population with colorectal cancer in The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center (MSKCC) databases. The specific aims of this study are i) to examine long-term survival characteristics of patients with mCRC; ii) to analyse and compare the clinical data of mCRC patients with different metastatic sites and patients with primary CRC and iii) to study the difference of gene mutation/expression in mCRC patients at different metastatic sites.
Materials and Methods
I Clinical Cohorts
The whole sequencing and clinical data of 2368 samples of colorectal cancer patients were obtained: Metastatic Colorectal Cancer (MSKCC, Cancer Cell 2018) 1134 samples, Colorectal Adenocarcinoma (TCGA, PanCancer Atlas) 594 samples, Colorectal Adenocarcinoma (TCGA, Firehouse Legacy) 640 samples ( Link) [24-27]. The patient cohort included 2329 incident cases diagnosed between 1995 and 2018.
II Study Design
We divided this study into four parts. In the first part, we compared clinical properties between Group B: metastatic colorectal cancer (mCRC) patients and Group A: non-metastatic primary colorectal cancer (pCRC) patients. In the second part, we analysed the clinical properties between mCRC patients with single-site tumor metastasis (Group C: liver, Group D: lung, Group E: lymph node, and Group F: peritoneum) and Group A: pCRC patients. We also focused on genomic changes in those with single-site tumor metastasis. In the third part, we explored and compared the gene mutations that were significantly correlated to the survival rate of CRC patients. Additional groups were as follows. Group G: mCRC-liver metastasis-KRAS mut patients; Group H: mCRC-liver metastasis-KRAS wild patients; Group I: mCRC-liver metastasis-TCF7L2 mut patients; Group J: mCRC-liver metastasis-TCF7L2 wild patients; Group K: mCRC-lung metastasis-APC mut patients; Group L: mCRC-lung metastasis-APC wild patients; Group M: mCRC-non-regional lymph node metastasis-APC mut patients; Group N: mCRC-non-regional lymph node metastasis- APC wild patients; Group O: mCRC-peritoneum metastasis-BRAF mut patients; Group P: mCRC- peritoneum metastasis-BRAF wild patients. Finally, in the fourth part, we used COX proportional hazards regression model to analyse which factors significantly contribute to the survival rate of patient in Group C, Group D, Group E, and Group F individually.
III Statistical Analysis
The data online were used to analyse without change. Frequency data was analysed by Chi-squared Test. Measurement Data (Quantitative) was analysed by Kruskal Wallis Test. Survival was illustrated by the Kaplan-Meier curves, with P value determined by Log-rank Test. Hazard’s ratio (HR) was determined through the univariate and multivariate COX proportional hazards regression model. Data analysis were performed using SPSS20.0(SPSS Inc. Chicago, IL).
Results
I Comparison of Clinical Characteristics Between Patients with Metastatic Colorectal Cancer (mCRC) and Primary Colorectal Cancer Patients Without Metastasis (pCRC)
Overall, our analysis included a total of 1133 patients diagnosed with pCRC (1137 samples) and 979 patients with mCRC (1011 samples), among which 565 (26.8%) were younger than 50-year-old, 913 (43.2%) were between 50 and 69-year-old (middle-aged), and 634 (30.0%) were older than 69-year-old (older-aged). More detailed information on the grouping is provided in (Table 1).
Table 1: Effect of gene mutation on survival rate of CRC patients with single-site tumor metastasis (liver, lung, lymph node, and peritoneum). To reduce statistical bias, patients with gene mutation rate >=10% and cells with the number of patients >=7 were selected. “----“ means patients with gene mutation rate <10% or the number of patients <7.
|
Mutation contribute to overall survival |
Only liver metastasis |
Only lung metastasis |
Only lymph metastasis |
Only peritoneum metastasis |
|
APC mutation |
No |
Yes* |
Yes* |
No |
|
TP53 mutation |
No |
No |
No |
No |
|
KRAS mutation |
Yes* |
No |
No |
No |
|
PIK3CA mutation |
No |
No |
No |
No |
|
SMAD4 mutation |
No |
---- |
---- |
No |
|
TCF7L2 mutation |
Yes* |
---- |
No |
---- |
|
ATM mutation |
---- |
No |
---- |
---- |
|
AMER1 mutation |
---- |
No |
---- |
---- |
|
ERBB4 mutation |
---- |
---- |
---- |
No |
|
NOTCH3 mutation |
---- |
---- |
---- |
No |
|
BRAF mutation |
---- |
---- |
---- |
Yes* |
Yes* Means P<0.05 mutation type gene compared with wild type gene.
First, we compared the impact of CRC metastases on the study population's overall survival (OS). The results indicated that the 10-year survival rate of patients with mCRC (group B) is significantly lower than that of patients with pCRC. There were 158 pCRC patients and 346 mCRC dead at the end of the study. The percentage of deaths was 13.9% (158/1,133) and 35.3% (346/979) in the pCRC and mCRC groups, respectively. The median survival for the mCRC group was 68.13 months. Next, we analysed the mutation frequency of the two groups. Only APC and TP53 gene mutations were significantly more frequent in patients with mCRC than in those with pCRC. However, the mutations of other genes including TTN, PIK3CA, MUC16, SYNE1, FBXW7, FAT4, and FLG in the metastatic foci of mCRC patients were reduced (Figure 1B, left panel). Copy number analysis indicated that only CDKN1B and MET genes were found to be amplified more frequently in metastatic lesions than in patients without metastasis and that deletion of RBFOX1, WWOX, CCSER1, MACROD2 and amplification of ASXL1, BCL2L1, TSPY26P, PLAGL2 were significantly reduced in metastatic foci (Figure 1B, right panel).
Figure 1: Comparison of clinical properties among metastatic colorectal cancer (mCRC) and non-metastatic primary colorectal cancer (pCRC) patients. A) overall survival; B) the mutation frequency (left panel) and copy number analysis (right panel); C) microsatellite instability (MSI) status; D) tumor sample histology analysis; E) primary tumor location analysis; F) Mutation Count; G) age at diagnosis; H) sex number samples. Asterisks indicate significant differences.
Tumor sample histology analysis suggests that tumors with conventional histological type are more likely to metastasize, while medullary carcinoma hardly metastasizes (Figure 1C). Mutation Count (log2) reveals that the number of gene mutations in mCRC patients is less than that in pCRC patients (Figure 1D). With regard to age at diagnosis, the average age of colon cancer patients without metastasis is 60-year-old, while that of colon cancer patients with metastasis is less than 60-year-old (Figure 1E), implying that we should pay more attention to the tumor metastasis tendency of young patients, increase examination methods in time, or be more radical in treatment strategy. Of note, primary tumor location analysis demonstrates that cancers originating from left colon are more prone to metastasis than cancers originating from right colon (Figure 1F). An intriguing finding of this study is that microsatellite instability (MSI) status in mCRC cancer patients is less than that in pCRC patients (Figure 1G). In addition, the gender is not a factor affecting the metastasis of colorectal cancer (Figure 1H). The statistical results of (Figures 1C-1H) are shown in (Table 2).
Table 2: The statistical results of (Figures 1C-1H and Figures 3A-3E).
|
|
Statistical Test |
p-Value |
|
Figure 1C Tumor Sample Histology |
Chi-squared Test |
P<0.01 |
|
Figure 1D Mutation Count |
Kruskal Wallis Test |
P<0.01 |
|
Figure 1E Age at Diagnosis |
Kruskal Wallis Test |
P<0.01 |
|
Figure 1F Primary Tumor Location |
Chi-squared Test |
P<0.01 |
|
Figure 1G MSI Status |
Chi-squared Test |
P<0.01 |
|
Figure 1H Sex |
Chi-squared Test |
P=0.969 |
|
Figure 3A MSI Status |
Chi-squared Test |
P<0.01 |
|
Figure 3B Tumor Sample Histology |
Chi-squared Test |
P<0.01 |
|
Figure 3C Primary Tumor Location |
Chi-squared Test |
P<0.01 |
|
Figure 3D Sex |
Chi-squared Test |
P=0.413 |
|
Figure 3E Age at Diagnosis |
Kruskal Wallis Test |
P<0.01 |
II Survival Rate Comparison Between mCRC Patients with Single-Site Tumor Metastasis and pCRC Patients, As Well as The Survival Rate Comparison Among mCRC Patients with Different Metastatic Foci
We next analysed survival discrepancies between mCRC patients with single-site tumor metastasis (liver, lung, lymph node, or peritoneum) and pCRC patients, thus focusing on the role of metastatic sites in mCRC. As shown in (Figures 2A & 2B), there is no statistical difference between the survival rates of mCRC patients with liver-only (mCRC-liver) or lung-only metastasis (mCRC-lung) and those of pCRC patients, suggesting that liver or lung metastasis does not reduce the survival rate of colon cancer patients. Strikingly, the survival rate of mCRC patients with lymph node metastasis is statistically different from that of pCRC patients (Figure 2C). It is worth noting that the lymph node metastasis here refers not to regional lymph node metastasis, but to non-regional lymph node metastasis. These results demonstrate that non-regional lymph node (NRLN) metastasis can significantly reduce the survival rate and is an unfavourable factor for mCRC patients. The clinical significance of this finding lies in that the resection scope should include non-regional lymph nodes during surgical treatment, and that detection of resected non-regional lymph nodes will help predict the risk of patients’ death, i.e., if the resected non-regional lymph nodes are positive, it indicates that the mortality rate of the patient will be increased compared with the patients with negative non-regional lymph nodes. In addition, we also demonstrated that peritoneal metastasis can significantly decrease the survival rate of mCRC patients and is another unfavourable factor (Figure 2D). This result suggests that we should pay more attention to the exclusion of peritoneal metastasis during preoperative examination and postoperative follow-up.
Figure 2: Comparison of survival rates between mCRC patients with single-site tumor metastasis and patients with pCRC, and comparison of survival rates among mCRC patients with different metastatic foci. A) mCRC patients with liver-only metastasis versus pCRC patients, B) mCRC patients with lung-only metastasis versus pCRC patients, C) mCRC patients with lymph node metastasis versus pCRC patients, D) mCRC patients with peritoneal metastasis versus pCRC patients, E) mCRC patients with non-regional lymph node only metastasis versus patients with liver only metastasis, F) mCRC patients with lung metastasis versus liver metastasis, G) non-regional lymph node metastasis versus lung metastasis, H) comparison and summary of median survival between each group.
Next, we examined the effect of different metastatic sites on survival rate of mCRC patients. The survival rate of mCRC patients with non-regional lymph node-only metastasis (mCRC-NRLN) (Figure 2E) or peritoneal-only metastasis (mCRC-peritoneum) (Figure S1) is lower than that of patients with mCRC-liver. Moreover, the survival rate of patients with mCRC-peritoneum is lower than that of patients with mCRC-lung (Figure S2). There was no statistical difference in survival rate between mCRC patients with lung metastasis and with liver metastasis (Figure 2F), between NRLN metastasis and lung metastasis (Figure 2G) or between NRLN metastasis and peritoneal metastasis (Figure S3). Taken together, one of the novel findings of this study is that liver or lung metastasis of mCRC patients does not affect the overall survival rate (Median Survival: 132 and 72.47 months, respectively), while peritoneal metastasis has the greatest impact on survival rate (Median Survival: 31.67 months) (Figure 2H).
III Comparison of Clinical Characteristics and Mutation between mCRC Patients with Specific Single-Site Tumor Metastasis and pCRC
Next, we further analysed the clinical characteristics of specific metastatic sites and their relationship with gene mutations. As shown in (Figure 3A), MSI status analysis suggests that patients with colorectal cancer accompanied by metastases have fewer microsatellite instable foci than patients with pCRC, of which mCRC-lung patients have the least MSI, followed by patients with mCRC-liver, patients with mCRC-RNLN and mCRC-peritoneum. Tumor sample histology analysis indicates that liver metastasis of colorectal cancer is more likely to occur in conventional histological types of tumors, peritoneal metastasis is more likely to occur in signet ring cell carcinoma, and medullary carcinoma hardly ever metastasizes (Figure 3B). Intriguingly, primary tumor location analysis shows that compared with cancers originating from the right colon, cancers originating from the left colon are most likely to metastasize to lungs, followed by liver and non-regional lymph nodes respectively. However, the metastatic rate of cancers originating from the right colon is relatively low (Figure 3C). Gender analysis indicates that there is no gender difference in tumor metastases of colorectal cancer patients (Figure 3D). Age at Diagnosis analysis showed that the average age of colon cancer patients is lowest, about 50-year-old, in those with lymph node metastasis is, followed by liver metastasis and peritoneal metastasis, and lastly lung metastasis, however the average age for all patients with metastasis was less than 60-year-old (Figure 3E). The statistical results of (Figures 3A-3E) shows in (Table 2).
Figure 3: Comparison of clinical features and mutation between mCRC patients with single-site tumor metastasis and pCRC patients. A) MSI status, B) tumor sample histology analysis, C) primary tumor location analysis, D) sex number samples, E) age at diagnosis, F) the mutation frequency analysis, G) copy number alteration frequency analysis.
These novel findings suggest that clinical oncologists should pay more attention to the tendency of lymph node metastasis in young CRC patients, increase examination methods in time, or pay closer attention to the dissection of non-regional lymph nodes during surgical treatment. In addition, the average age of mCRC patients with lung metastases is relatively older, which also suggests that we should pay more attention to the screening of lung metastases for older patients. In order to find additional predictive biomarkers or molecular signatures, we performed mutation frequency and copy-number analysis. We selected the genes with the top 10 mutation rates in single-site tumor metastases (liver, lung, lymph node, peritoneum) for analysis. As indicated in (Figure 3F), the APC mutation rate in liver and lung metastasis is higher than that in patients with pCRC. The mutation rate of TP53 in almost all metastatic foci is higher than that in patients without tumor metastasis. KRAS mutation rate increases significantly in mCRC patients with lung metastasis, and the proportion of ERBB4, NOTCH3 and BRAF mutations increases in patients with peritoneal metastasis. The trend of gene copy-number is the same as that of (Figure 1).
As shown in (Figure 3G), the frequency of CDKN1B gene amplification in liver, lymph node and peritoneal metastases is higher than that in non-metastatic patients, and the frequency of MET gene amplification in liver, lung and lymph node metastases is higher than that in non-metastatic patients. The deletions of RBFOX1, WWOX, CCSER1, and MACROD2 in all metastatic foci are significantly reduced. The amplification of BCL2L1 in lymph node metastasis is higher than that in tumor samples of patients with pCRC without metastasis, but the number of patients (4) is too small to draw strong conclusions, only 4. Mutation Count (log2) analysis reveals that the number of gene mutations in patients with mCRC is less than that in pCRC patients. However, for different types of metastases, there is no difference in the number of mutations between liver, lung, lymph node and peritoneal metastases (Figure S4). Previous studies have shown that for mCRC patients, the mutation genes in the primary foci and metastatic foci are exactly the same [28]. Based on our study and this previous evidence, there are likely more mutations in the primary foci of pCRC patients and compared to the primary foci of mCRC patients, suggesting that mutations in the primary foci could be used to predict and evaluate the possibility of CRC metastasis.
IV Gene Mutations That Significantly Affect the Survival Rate of CRC Patients with Different Single-Site Metastases
In order to further determine the role of gene mutation in CRC metastasis, we analysed the effect of gene mutation on survival rate of CRC patients with single-site tumor metastasis. All metastatic foci of CRC patients have roughly the same range of mutated genes. However, the same gene mutation in different single-site tumor metastases (liver, lung, lymph nodes, and peritoneum) has different effects on survival rate. Specifically, as shown in (Table 1), the KRAS and TCF7L2 gene mutation have a significant impact on the survival rate of mCRC patients accompanied by liver only metastasis, APC gene mutation has a significant effect on the survival rate of patients with mCRC-lung or mCRC-NRLN, and in mCRC patients accompanied by peritoneal only metastasis, BRAF gene mutation has a significant impact on their survival rates. More details are shown in (Table 1), * means P<0.05 compared with wild type gene. The above results also suggest that different mutant genes have distinct effects on the survival rate of mCRC patients with different single-site tumor metastases, implying that gene mutations have relative specificity for the metastatic site of CRC.
In order to further understand the specific impact of these gene mutations on the survival rate of CRC patients with single-site metastasis, we compared survival rates between patients with gene mutations and those with wild-type genes. In patients with mCRC-liver, the survival rate of patients with the KRAS gene mutation is lower than that of patients with the wild-type KRAS gene, suggesting that KRAS mutation may be associated with reduced survival rate of patients with mCRC-liver and be used as an indicator for predicting survival rate of such patients (Figure 4A). Further analysis shows that TCF7L2 gene mutation may be associated with increased survival rate of patients with mCRC-liver (Figure 4B), APC gene mutation is associated with increased survival rate of patients with mCRC-lung (Figure 4C) and mCRC-NRLN (Figure 4D), and BRAF gene mutation is related to reduced survival rate of patients with mCRC-peritoneum (Figure 4E). The median survival for all these patients is summarized in (Figure 4F).
Figure 4: Gene mutations that dramatically affect the survival rate of CRC patients with distinct single-site metastases. A) mCRC-liver patients with KRAS mutation versus those with KRAS WT, B) mCRC-liver patients with TCF7L2 mutation versus TCF7L2 WT, C) mCRC-lung patients with APC mutation versus APC WT, D) mCRC-NRLN patients with APC mutation versus APC WT, E) mCRC-peritoneum patients with BRAF mutation versus BRAF WT, F) Comparison of median months survival.
Our research has positive guiding significance for clinical management of patients with colorectal cancer. For mCRC patients with liver metastases, the prognosis can be predicted by mutation of KRAS and TCF7L2 genes. KRAS mutation may predict increased mortality of mCRC-liver patients, while TCF7L2 mutation predicts reduced mortality. The mutation of APC gene can be used to predict the mortality rate of patients with mCRC-lung or mCRC-NRLN, i.e., APC mutation indicates reduced mortality of these patients. In addition, for mCRC patients with peritoneal metastasis, the mortality can be predicted by mutation of BRAF gene. If these patients have BRAF mutations, we can predict the prognosis will be poor.
V Cox Proportional Hazards Regression Model Analysis on Survival Rate of mCRC Patients with Liver-Only Metastasis
We further comprehensively analysed the factors affecting the survival rate of patients with mCRC-liver. Univariate COX Risk Model Analysis (Tables 3 & 4) suggests these factors include KRAS_status, Fraction_Genome_Altered, Chemo_Exposure_of_sequenced_specimen (Whether receive chemotherapy), Patient_Tumor_Grade, and Time_from_Met_Dx_to_Sequencing. Specifically, KRAS gene mutation is an unfavourable factor for prognosis of mCRC-liver patients. Compared with KRAS wild type (WT) mCRC-liver patients, mCRC-liver patients with KRAS gene mutation have 1.308 times higher risk of death. With the increase of the fraction genome altered, the risk of death in mCRC-liver patients decreased. Compared with patients who do not receive chemotherapy, mCRC-liver patients receiving chemotherapy have a lower risk of death. The risk of death for mCRC-liver patients with intermediate differentiation and intermediate-low differentiation is lower than that of mCRC-liver patients with low differentiation. mCRC-liver patients receiving longer-term chemotherapy have a lower risk of death than receiving shorter-term chemotherapy.
Table 3: Baseline characteristics of mCRC patients with liver only metastasis.
|
Variable |
Only liver metastasis(n=520) |
|
KRAS_status (%) |
|
|
Mut |
326(62.7%) |
|
Wild |
194(37.3%) |
|
TCF7L2_status (%) |
|
|
Mut |
476(91.5%) |
|
Wild |
44(8.5%) |
|
Age_at_Diagnosis,median(range),years |
53.94(24-83) |
|
Sex, n(%) |
|
|
Male |
299(57.5%) |
|
Female |
221(42.5%) |
|
Fraction_Genome_Altered |
0.21(0-0.78) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
204(39.2%) |
|
Yes |
316(60.8%) |
|
Molecular_Subtype(%) |
|
|
MSS |
507(97.5%) |
|
MSI |
11(2.1%) |
|
Unknow |
2(0.4%) |
|
MSI_Score |
1.59(0-47.7) |
|
Mutation_Count |
8.85(0-249) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
34(6.5%) |
|
Mod-Poorly-Diff |
36(6.9%) |
|
Mod_Diff |
326(62.7%) |
|
Unknow |
124(23.8%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
40(7.7%) |
|
Cecum |
51(9.8%) |
|
Descending Colon |
35(6.7%) |
|
Hepatic Flexure |
13(2.5%) |
|
Rectosigmoid |
61(11.7%) |
|
Rectum |
111(21.3%) |
|
Sigmoid Colon |
162(31.2%) |
|
Splenic Flexure |
11(2.1%) |
|
Transverse Colon |
28(5.4%) |
|
Unknow |
8(1.5%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
390(75%) |
|
Right |
124(23.8%) |
|
Unknow |
6(1.2%) |
|
Sample_Type, n(%) |
|
|
Primary |
209(40.2%) |
|
Metastasis |
311(59.8%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
32.39(-7.53-270.53) |
|
Time_to_Metastasis, median (range),month |
4.69(0-120.00) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
326(62.7%) |
|
Conventional_With_Mucinous_Component |
30(5.8%) |
|
Mucinous |
6(1.2%) |
|
PDC |
33(6.3%) |
|
Unknow |
125(24%) |
|
Overall_Survival_Months, median(range),month |
47.26(0.57-292.93) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
151(29%) |
|
Living |
369(71%) |
Table 4: COX hazard analysis of overall survival for mCRC patients with liver only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
|
P |
HR |
95%CI |
|
|||
|
down |
upper |
down |
upper |
|
||||||
|
KRAS_status |
0.014 |
1.308 |
1.056 |
1.620 |
0.863 |
0.973 |
0.716 |
1.323 |
|
|
|
TCF7L2_status |
0.951 |
0.989 |
0.705 |
1.388 |
|
0.326 |
1.239 |
0.808 |
1.901 |
|
|
Age_at_Diagnosis |
0.873 |
0.999 |
0.989 |
1.009 |
|
0.779 |
1.002 |
0.989 |
1.014 |
|
|
Sex |
0.200 |
0.872 |
0.708 |
1.075 |
|
0.037 |
1.334 |
1.018 |
1.749 |
|
|
Fraction_Genome_Altered |
0.002 |
0.348 |
0.179 |
0.676 |
|
0.343 |
0.594 |
0.203 |
1.743 |
|
|
Chemo_Exposure_of_sequenced_specimen |
≤0.001 |
0.444 |
0.358 |
0.550 |
|
0.024 |
1.454 |
1.050 |
2.015 |
|
|
Molecular_Subtype |
0.607 |
1.180 |
0.628 |
2.217 |
|
0.892 |
0.641 |
0.001 |
389.262 |
|
|
MSI_Score |
0.728 |
0.996 |
0.977 |
1.017 |
|
0.944 |
1.006 |
0.851 |
1.189 |
|
|
Mutation_Count |
0.183 |
1.003 |
0.998 |
1.008 |
|
0.875 |
1.004 |
0.950 |
1.062 |
|
|
Patient_Tumor_Grade |
0.046 |
0.802 |
0.645 |
0.997 |
|
0.426 |
0.846 |
0.560 |
1.278 |
|
|
Primary_Tumor_Site |
0.447 |
0.981 |
0.934 |
1.030 |
|
0.137 |
1.064 |
0.981 |
1.154 |
|
|
Primary_Tumor_Location |
0.041 |
1.300 |
1.011 |
1.673 |
|
0.914 |
1.025 |
0.656 |
1.601 |
|
|
Sample_Type |
≤0.001 |
0.446 |
0.357 |
0.555 |
|
0.548 |
0.903 |
0.647 |
1.260 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.822 |
0.806 |
0.838 |
|
≤0.001 |
0.811 |
0.792 |
0.831 |
|
|
Time_to_Metastasis |
0.907 |
1.000 |
0.992 |
1.007 |
|
0.732 |
1.002 |
0.992 |
1.012 |
|
|
Tumor_Sample_Histology |
0.179 |
1.111 |
0.953 |
1.294 |
|
0.891 |
1.020 |
0.768 |
1.355 |
|
Interestingly, multivariate cox risk model analysis yielded unexpected results. Compared with male mCRC-liver patients, female mCRC-liver patients have a higher risk of death (approximately 1.334 times). The death risk of mCRC-liver patients receiving chemotherapy is 1.454 times higher than that of mCRC-liver patients without chemotherapy. It is worth noting that this multivariate COX analysis conclusion is contrary to the previous univariate COX analysis conclusion, nonetheless the multivariate COX analysis conclusion is more important. Although chemotherapy itself is an unfavourable factor, the time of receiving chemotherapy is a favourable prognostic factor for mCRC-liver patients. mCRC-liver patients receiving longer-term chemotherapy have a lower risk of death.
VI Cox Proportional Hazards Regression Model Analysis on Survival Rate of mCRC Patients with Other Single-Site Metastases
Univariate COX risk model analysis indicates that factors affecting survival rate of colorectal cancer patients with mCRC-lung are Fraction Genome Altered and Time from metastasis diagnosis Met Dx to Sequencing (Tables 5 & 6). Specifically, mCRC-lung patients with higher fraction genome altered or receiving longer-term chemotherapy have a lower risk of death. Multivariate COX risk model analysis demonstrates that factors affecting survival rate of mCRC-lung patients include age at diagnosis, primary tumor site and time from met dx to sequencing. Age at diagnosis is an unfavourable factor affecting the survival rate of patients with mCRC-lung. The older the patients at the time of diagnosis, the higher the risk of death. Intriguingly, compared with mCRC-lung patients whose primary tumor site is ascending colon, mCRC-lung patients whose primary tumor site is transverse colon or splenic flexure are less at risk of death. However, because there are so many groups of primary tumor sites that the number of patients in each group is very small, we think this result is not reliable and is a false positive result. Lastly, the time of receiving chemotherapy is a favourable factor for prognosis of mCRC-lung patients. mCRC-lung patients receiving longer-term chemotherapy have a lower risk of death.
Table 5: Baseline characteristics of mCRC patients with lung-only metastasis.
|
Variable
|
Only lung metastasis(n=63)
|
|
APC_status (%) |
|
|
Mut |
50(79.4%) |
|
Wild |
13(20.6%) |
|
Age_at_Diagnosis,median(range),years |
56.69(29-84) |
|
Sex, n(%) |
|
|
Male |
33(52.4%) |
|
Female |
30(47.6%) |
|
Fraction_Genome_Altered |
0.1948(0-0.57) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
24(38.1%) |
|
Yes |
39(61.9%) |
|
Molecular_Subtype(%) |
|
|
MSS |
62(98.4%) |
|
MSI |
1(1.6%) |
|
MSI_Score |
1.59(0-47.7) |
|
Mutation_Count |
7.11(1-44) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
2(3.2%) |
|
Mod-Poorly-Diff |
4(6.3%) |
|
Mod_Diff |
41(65.1%) |
|
Unknow |
16(25.4%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
1(1.6%) |
|
Cecum |
5(7.9%) |
|
Descending Colon |
3(4.8%) |
|
Rectosigmoid |
10(15.9%) |
|
Rectum |
34(54.0%) |
|
Sigmoid Colon |
9(14.3%) |
|
Transverse Colon |
1(1.6%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
57(90.5%) |
|
Right |
6(9.5%) |
|
Sample_Type, n(%) |
|
|
Primary |
26(41.3%) |
|
Metastasis |
37(58.7%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
21.25(0.90-130.67) |
|
Time_to_Metastasis, median (range),month |
18.61(0-68.20) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
38(60.3%) |
|
Conventional_With_Mucinous_Component |
5(7.9%) |
|
Mucinous |
2(3.2%) |
|
PDC |
2(3.2%) |
|
Unknow |
16(25.4%) |
|
Overall_Survival_Months, median(range),month |
36.16(1.3-138.03) |
|
Overall_Survival_Status (%) |
|
|
Deceased |
20(31.7%) |
|
Living
|
43(68.3%)
|
Table 6: COX hazard analysis of overall survival for mCRC patients with lung only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||
|
down |
upper |
|
down |
upper |
||||||
|
APC _status |
0.885 |
0.941 |
0.415 |
2.134 |
0.065 |
0.255 |
0.059 |
1.090 |
||
|
Age_at_Diagnosis |
0.420 |
1.010 |
0.985 |
1.036 |
|
0.011 |
1.055 |
1.012 |
1.099 |
|
|
Sex |
0.398 |
0.772 |
0.423 |
1.408 |
|
0.669 |
1.283 |
0.409 |
4.019 |
|
|
Fraction_Genome_Altered |
0.027 |
0.103 |
0.014 |
0.769 |
|
0.583 |
3.272 |
0.047 |
226.317 |
|
|
Chemo_Exposure_of_sequenced_specimen |
0.394 |
0.767 |
0.416 |
1.413 |
|
0.154 |
2.305 |
0.732 |
7.258 |
|
|
Molecular_Subtype |
0.945 |
1.072 |
0.146 |
7.886 |
|
0.861 |
0.080 |
0.000 |
1.598e11 |
|
|
MSI_Score |
0.608 |
0.975 |
0.884 |
1.075 |
|
0.718 |
1.186 |
0.471 |
2.984 |
|
|
Mutation_Count |
0.897 |
0.996 |
0.945 |
1.051 |
|
0.598 |
0.935 |
0.729 |
1.199 |
|
|
Patient_Tumor_Grade |
0.613 |
1.388 |
0.389 |
4.952 |
|
0.720 |
0.762 |
0.173 |
3.358 |
|
|
Primary_Tumor_Site |
0.262 |
0.898 |
0.745 |
1.083 |
|
0.021 |
0.412 |
0.194 |
0.875 |
|
|
Primary_Tumor_Location |
0.756 |
1.179 |
0.417 |
3.327 |
|
0.071 |
0.045 |
0.002 |
1.305 |
|
|
Sample_Type |
0.173 |
0.648 |
0.347 |
1.210 |
|
0.117 |
2.391 |
0.805 |
7.107 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.866 |
0.821 |
0.914 |
|
≤0.001 |
0.829 |
0.767 |
0.896 |
|
|
Time_to_Metastasis |
0.966 |
1.000 |
0.982 |
1.020 |
|
0.698 |
0.991 |
0.944 |
1.039 |
|
|
Tumor_Sample_Histology |
0.641 |
1.149 |
0.642 |
2.056 |
|
0.900 |
0.948 |
0.414 |
2.174 |
|
Table 7: Baseline characteristics of mCRC patients with lymph-only metastasis.
|
Variable |
Only lymph metastasis (n=40) |
|
APC_status (%) |
|
|
Mut |
29(72.5%) |
|
Wild |
11(27.5%) |
|
Age_at_Diagnosis,median(range),years |
51.73(26-81) |
|
Sex, n(%) |
|
|
Male |
26(65%) |
|
Female |
14(35%) |
|
Fraction_Genome_Altered |
0.198(0-0.96) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
27(67.5%) |
|
Yes |
13(32.5%) |
|
Molecular_Subtype(%) |
|
|
MSS |
35(87.5%) |
|
MSI |
4(10%) |
|
Unknow |
1(2.5%) |
|
MSI_Score |
3.70(0-37.38) |
|
Mutation_Count |
16.08(1-159) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
4(10%) |
|
Mod-Poorly-Diff |
6(15%) |
|
Mod_Diff |
18(45%) |
|
Unknow |
12(30%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
3(7.5%) |
|
Cecum |
8(20.0%) |
|
Descending Colon |
2(5%) |
|
Rectosigmoid |
4(10%) |
|
Rectum |
12(30%) |
|
Sigmoid Colon |
9(22.5%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
28(70%) |
|
Right |
12(30%) |
|
Sample_Type, n(%) |
|
|
Primary |
32(80%) |
|
Metastasis |
8(20%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
15.5(-19.63-100.8) |
|
Time_to_Metastasis, median (range),month |
14.90(0-125.8) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
19(47.5%) |
|
Conventional_With_Mucinous_Component |
4(10%) |
|
Mucinous |
1(2.5%) |
|
PDC |
3(7.5%) |
|
Signet_Ring_Cell |
1(2.5%) |
|
Unknow |
12(30%) |
|
Overall_Survival_Months, median(range),month |
28.02(0.93-126) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
16(40%) |
|
Living |
24(60%) |
Because isolated non-regional lymph node metastasis (NRLN) in CRC patients is uncommon, little is known about the factors that affect the survival rate of such patients. Here, our univariate COX risk model analysis shows that factors affecting survival rate of patients with mCRC-NRLN are chemo exposure of sequenced specimen and time from Met Dx to sequencing (Tables 7 & 8). Specifically, chemotherapy and the time of receiving chemotherapy are favourable factors for prognosis of mCRC-NRLN patients. Interestingly, multivariate COX risk model analysis indicates that gene mutation count and the time of receiving chemotherapy are favourable factors for prognosis of mCRC-NRLN patients (Tables 7 & 8). In other words, mCRC-NRLN patients with higher gene mutation count have lower risk of death.
Table 8: COX hazard analysis of overall survival for mCRC patients with lymph-only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||
|
down |
upper |
|
down |
upper |
||||||
|
APC _status |
0.205 |
0.462 |
0.14 |
1.523 |
0.79 |
0.665 |
0.033 |
13.407 |
||
|
Age_at_Diagnosis |
0.67 |
1.01 |
0.966 |
1.055 |
|
0.331 |
0.943 |
0.838 |
1.062 |
|
|
Sex |
0.388 |
1.488 |
0.604 |
3.665 |
|
0.7 |
1.715 |
0.11 |
26.722 |
|
|
Fraction_Genome_Altered |
0.105 |
0.171 |
0.020 |
1.446 |
|
0.982 |
0.863 |
0 |
2.68*10^5 |
|
|
Chemo_Exposure_of_sequenced_specimen |
0.023 |
0.297 |
0.104 |
0.847 |
|
0.588 |
0.472 |
0.031 |
7.113 |
|
|
Molecular_Subtype |
0.161 |
2.47 |
0.697 |
8.75 |
|
0.969 |
2.7e22 |
0 |
-- |
|
|
MSI_Score |
0.173 |
1.037 |
0.984 |
1.094 |
|
0.737 |
0.686 |
0.076 |
6.173 |
|
|
Mutation_Count |
0.592 |
0.997 |
0.986 |
1.008 |
|
0.038 |
0.565 |
0.329 |
0.969 |
|
|
Patient_Tumor_Grade |
0.367 |
0.703 |
0.326 |
1.513 |
|
0.956 |
1.055 |
0.156 |
7.14 |
|
|
Primary_Tumor_Site |
0.982 |
0.998 |
0.823 |
1.210 |
|
0.973 |
1.02 |
0.332 |
3.134 |
|
|
Primary_Tumor_Location |
0.886 |
1.068 |
0.435 |
2.618 |
|
0.546 |
5.469 |
0.022 |
1356.578 |
|
|
Sample_Type |
0.091 |
0.382 |
0.125 |
1.165 |
|
0.854 |
0 |
0 |
1.16E+36 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.867 |
0.814 |
0.925 |
|
0.002 |
0.757 |
0.637 |
0.901 |
|
|
Time_to_Metastasis |
0.263 |
0.991 |
0.976 |
1.007 |
|
0.689 |
1.01 |
0.964 |
1.058 |
|
|
Tumor_Sample_Histology |
0.501 |
1.180 |
0.729 |
1.911 |
|
0.963 |
1.037 |
0.221 |
4.854 |
|
Lastly, we performed COX analysis to determine the factors affecting the survival of patients with mCRC-peritoneum. Univariate COX risk model analysis shows that there are no factors affecting the survival rate of mCRC-peritoneum patients. However, multivariate COX risk model analysis shows that Age at Diagnosis, Chemo Exposure of sequenced specimen, Molecular Subtype, Mutation Count, Primary Tumor Site and Sample Type all affect the survival rate of mCRC-peritoneum patients (Tables 9 & 10). Due to the small sample of mCRC-peritoneum patients, we consider these results unreliable.
Table 9: Baseline characteristics of colorectal cancer patients with peritoneum-only metastasis.
|
Variable |
Only peritoneum metastasis(n=52) |
|
BRAF_status (%) |
|
|
Mut |
8(15.4%) |
|
Wild |
44(84.6%) |
|
Age_at_Diagnosis,median(range),years |
54.52(13-80) |
|
Sex, n(%) |
|
|
Male |
28(53.8%) |
|
Female |
24(46.2%) |
|
Fraction_Genome_Altered |
0.133(0-0.63) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
32(61.5%) |
|
Yes |
20(38.5%) |
|
Molecular_Subtype(%) |
|
|
MSS |
41(78.8%) |
|
MSI |
11(21.2%) |
|
MSI_Score |
6.39(0-42.22) |
|
Mutation_Count |
15.75(2-90) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
9(17.3%) |
|
Mod-Poorly-Diff |
7(13.5%) |
|
Mod_Diff |
18(34.6%) |
|
Unknow |
18(34.6%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
6(11.5%) |
|
Cecum |
13(25%) |
|
Descending Colon |
4(7.7%) |
|
Rectosigmoid |
1(1.9%) |
|
Rectum |
5(9.6%) |
|
Sigmoid Colon |
13(25%) |
|
Hepatic Flexure |
4(7.7%) |
|
Splenic Flexure |
2(3.8%) |
|
Transverse Colon |
3(5.8%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
26(50%) |
|
Right |
26(50%) |
|
Sample_Type, n(%) |
|
|
Primary |
30(57.7%)) |
|
Metastasis |
22(42.3%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
-10.59(-1365.80-159.37) |
|
Time_to_Metastasis, median (range),month |
4.49(0-62.9) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
17(32.7%)) |
|
Conventional_With_Mucinous_Component |
7(13.5%) |
|
Mucinous |
1(1.9%) |
|
PDC |
5(9.6%) |
|
Signet_Ring_Cell |
3(5.8%) |
|
Overall_Survival_Months, median(range),month |
26.8(1.87-173.57) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
24(46.2%) |
|
Living |
28(53.8%) |
Table 10: COX hazard analysis of overall survival for mCRC patients with peritoneum-only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||||||
|
down |
upper |
|
down |
upper |
||||||||||
|
BRAF_status |
0.909 |
0.918 |
0.211 |
3.998 |
0.616 |
1.94 |
0.146 |
25.854 |
|
|||||
|
Age_at_Diagnosis |
0.414 |
0.989 |
0.963 |
1.016 |
|
0.001 |
0.884 |
0.821 |
0.951 |
|||||
|
Sex |
0.486 |
0.754 |
0.340 |
1.670 |
|
0.427 |
1.78 |
0.429 |
7.384 |
|||||
|
Fraction_Genome_Altered |
0.845 |
1.281 |
.108 |
15.265 |
|
0.365 |
7.989 |
0.089 |
713.333 |
|||||
|
Chemo_Exposure_of_sequenced_specimen |
0.225 |
0.597 |
0.259 |
1.374 |
|
0.015 |
22.192 |
1.815 |
271.313 |
|||||
|
Molecular_Subtype |
0.615 |
0.784 |
0.304 |
2.023 |
|
0.011 |
0 |
0 |
0.003 |
|||||
|
MSI_Score |
0.490 |
0.989 |
0.959 |
1.020 |
|
0.686 |
0.945 |
0.717 |
1.245 |
|||||
|
Mutation_Count |
0.321 |
0.992 |
0.975 |
1.008 |
|
0.004 |
1.668 |
1.177 |
2.364 |
|||||
|
Patient_Tumor_Grade |
0.192 |
0.699 |
0.408 |
1.197 |
|
0.305 |
2.408 |
0.45 |
12.885 |
|||||
|
Primary_Tumor_Site |
0.338 |
0.931 |
0.803 |
1.078 |
|
0.008 |
0.42 |
0.222 |
0.795 |
|||||
|
Primary_Tumor_Location |
0.557 |
1.257 |
.586 |
2.694 |
|
0.089 |
0.09 |
0.006 |
1.441 |
|||||
|
Sample_Type |
0.131 |
0.538 |
0.241 |
1.202 |
|
0.001 |
0.009 |
0 |
0.151 |
|||||
|
Time_from_Met_Dx_to_Sequencing |
0.081 |
0.999 |
0.998 |
1.000 |
|
0.239 |
0.997 |
0.993 |
1.002 |
|||||
|
Time_to_Metastasis |
0.593 |
1.008 |
0.978 |
1.039 |
|
0.915 |
0.994 |
0.896 |
1.103 |
|||||
|
Tumor_Sample_Histology |
0.188 |
1.227 |
0.905 |
1.663 |
|
0.303 |
1.721 |
0.613 |
4.832 |
|||||
Discussion
Recent advances in understanding metastasis at molecular and gene levels have aroused increasing interest in the epidemiology of metastatic colorectal cancer (mCRC). In the present study, we assessed the relationship between metastatic patterns and survival outcomes of CRC patients, and sought to identify some prognostic predictors using the whole sequencing and clinical data of 2368 samples (2329 CRC patients) obtained from (TCGA and MSKCC databases). Such a long follow-up period makes it a brilliant resource for studying short-term and long-term prognosis characteristics. Our results showed an important prognostic value of the distant metastasis sites at diagnosis and effect of specific gene mutation on mortality of mCRC patients with single site metastasis (Figure 5). Additionally, we have determined the independent prognostic factors of patients with liver-only metastasis and unanimously found that a longer period of chemotherapy is associated with survival of patients.
Figure 5: Schematic diagram summarizing factors closely related to the overall survival of CRC patients with different single site metastasis.
I Prognostic Value of Tumor Metastasis Sites on Survival Rate
Several studies have compared the survival rates of CRC patients with different metastatic sites, however, most research is based on single institutional experience with a fairly limited sample size or concentrated in a particular metastasis site [15, 18, 29, 30]. The analysis reported here makes use of a large number of patient groups, and thoroughly examines the influence of metastatic sites on survival outcomes by adjusting demographic and clinical variables that may affect survival rates. We report the following novel findings. i) Compared with non-metastatic CRC patients, the mortality rate of CRC patients with liver-only metastasis (mCRC-liver) did not increase. ii) The mortality rate of CRC patients with non-regional lymph node-only metastasis (mCRC-NRLN) was significantly higher than that of patients without metastasis. This finding suggests that during surgical treatment, the scope of resection should include non-regional lymph nodes, and their pathological examination results can be used to predict the risk of death. iii) The mortality rate of CRC patients with peritoneum-only metastasis (mCRC-peritoneum) was significantly higher than that of patients without metastasis, suggesting that we should pay more attention to the exclusion of peritoneal metastasis during preoperative examination and postoperative follow-up. iv) It has been reported in the literatures in small patient populations that even after multivariate adjustment, patients with lung-only metastasis (mCRC-lung) have a survival advantage over mCRC-liver [20, 31-33]. Inconsistent with this conclusion, we found that there is no significant difference in survival rate between patients with mCRC-lung and those with mCRC-liver. However, from (Figure 2), we can see that the median survival of patients with mCRC-liver is 132 months, while the median survival of patients with mCRC-lung is 72.47 months, with obvious difference in value. More research is needed in a larger patient population to elucidate this issue. v) The survival rate of mCRC-NRLN patients is not statistically different from that of mCRC-lung patients, but it is lower than that of mCRC-liver patients. This difference in statistical significance may again be related to differences in group sizes. The next step is to further increase the sample size of patients with mCRC-lung for further analysis. In addition, we did not include CRC patients with two or more metastatic sites at the same time in this study, due to the insufficient number of cases.
II Prognostic Significance of Genetic Mutations in CRC patients
It is now generally believed that sporadic CRC is usually caused by precancerous lesions via activation of oncogenes (KRAS and BRAF), inactivation of tumor suppressor genes (such as APC, p16, p53 and DCC) and mismatch repair genes (MLH1 and MSH2), as well as PMS2 and hMSH6 to a lesser extent [34]. In addition, previous studies have clearly indicated that APC, TP53, KRAS and other gene mutations play a role in the occurrence, development and metastasis of CRC [35-40]. The genetic alterations might influence the survival of the patients with CRC. Next, we sought to determine the correlation between mutation of specific genes and mortality of CRC patients to find more accurate indicators for predicting CRC mortality, so as to provide guidance for clinical practice. We demonstrated that for CRC patients who already have liver metastasis, mortality can be predicted by mutations of KRAS and TCF7L2 genes. KRAS mutation may indicate increased while TCF7L2 mutation indicates reduced mortality of patients with mCRC-liver. For patients with mCRC-lung and mCRC-NRLN, chance of mortality can be predicted by mutation of APC gene. APC mutation reduces likelihood of mortality in these patients. Furthermore, the mutation of BRAF gene can be used to predict the chance of mortality of mCRC-peritoneum patients. BRAF mutation increases the likelihood of mortality in these patients.
In the COX univariate analysis (Tables 4, 6, 8 & 10), we only observed that KRAS gene mutation is associated with the survival rate of mCRC-liver patients. No gene mutation other than KRAS has been observed to be correlated with the survival rate of mCRC patients with single-site metastasis. Notably, the effect of KRAS gene mutation on survival rate of mCRC-liver patients was not significant in COX multivariate analysis. Surprisingly, we found in COX multivariate analysis that male and female sex does affect the survival rate of mCRC-liver patients. Why is this happening? The reason may be that males and females have different gene mutations, so the correlation between KRAS gene mutation and survival rate is no longer important compared with the influence of gender on survival rate. The specific mechanism needs further study.
III The Influence of Other Factors on Survival Rate
As mentioned above, through multivariate COX risk model analysis, we found for the first time that gender could affect the survival rate of mCRC-liver patients. Female gender is an unfavourable factor for prognosis of mCRC-liver patients. Compared with male patients with mCRC-liver, female patients with mCRC-liver have a higher risk of death, which is 1.334 times that of male mCRC-liver patients. Moreover, we demonstrated that age would affect the survival rate of patients with mCRC-lung and that the number of tumor gene mutations affect the survival rate of mCRC-NRLN patients. Lastly, our multivariate COX risk model analysis indicates that chemo exposure of specimens can also affect the survival rate of mCRC-liver patients. Receiving chemotherapy is an unfavourable factor for prognosis of mCRC-liver patients. Compared with patients who do not receive chemotherapy, mCRC-liver patients who receive chemotherapy have a higher risk of death (1.454 times, p<0.05). Intriguingly, although receiving chemotherapy is an unfavourable factor for the prognosis of mCRC-liver patients, the length of chemotherapy treatment is an advantageous prognosis factor (Table 4). These results may seem contradictory, but they are not. There is more or less damage to liver function in mCRC-liver patients and the side effects of the chemotherapy drugs with conventional dosages are relatively large for them. Side effects of chemotherapy drugs may be one of the risk factors leading to death of these patients. Therefore, we suggest that for patients with mCRC-liver, it is necessary to reduce the side effects of chemotherapy drugs, reduce the use of chemotherapy drugs with greater liver toxicity, and appropriately extend the chemotherapy time for these patients, so as to obtain a better survival expectation.
In addition, we also found that some pathological factors are related to the metastatic tendency of CRC patients. For example, tumors with conventional histological type are more likely to metastasize, while medullary carcinoma hardly metastasizes (Figure 1C), and cancers originating from left colon are more prone to metastasis than cancers originating from right colon (Figure 1F). Therefore, in addition to well-established clinico-pathological factors, histological classification and localization of primary tumor can be used to predict metastasis, disease recurrence, and clinical outcome of CRC patients.
Conclusion
In summary, this study depicts the long-term survival features of a group of CRC patients and the comprehensive relationship between baseline variables and long-term patient outcomes. Overall, our findings expand the scope of information about patient outcomes and long-term risk markers, offer a new perspective that may be helpful for future research, and have positive guiding significance for clinical management of CRC patients. A staging system that comprehensively considers factors such as metastatic sites, gender of patients, selection of chemotherapy drugs, length of chemotherapy treatment, specific gene mutation, etc., can better stratify the treatment risks of CRC patients and more accurately predict their survival.
Availability of Data and Materials
The datasets used for the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
None.
Author Contributions
WD and YW designed the work. WD, KS, MS and YW analysed and interpreted the patient data. YW, KS and JV were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Acknowledgment
This work was supported in part by NIH-NIMHD U54MD007598 and U54CA143931, NIH/NCI1U54CA14393, U56 CA101599-01; Department-of-Defense Breast Cancer Research Programme grant BC043180, NIH/NCATS CTSI UL1TR000124 to J.V. Vadgama; Accelerating Excellence in Translational Science Pilot Grants G0812D05, NIH/NCI SC1CA200517 and 9 SC1 GM135050-05 to Y. Wu; This research was partially supported by NIH National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR001881, NIH-NIMHD U54MD007598, and NIMHD S21 MD000103. NIH-NIMHD, NIH/NCATS and NIH/NCI have had no involvement in the study design, collection, analysis and interpretation of data, the writing of the report, or the decision to submit the article for publication.
Consent for Publication
The content of this manuscript has not been previously published and is not under consideration for publication elsewhere.
Abbreviations
APC: Adenomatous Polyposis Coli
ASXL1: Additional Sex Combs Like 1
BCL2L1: Bcl-2-Like Protein 1
BRAF: B-Raf Proto-Oncogene
CCSER1: Coiled-Coil Serine Rich Protein 1
CDKN1B: Cyclin Dependent Kinase Inhibitor 1B
CRC: Colorectal Cancer
DCC: Deleted In Colorectal Carcinoma
ERBB4: Erb-B2 Receptor Tyrosine Kinase 4
FAT4: FAT Atypical Cadherin 4
FBXW7: F-Box/WD Repeat-Containing Protein 7
FLG: Filaggrin
HR: Hazard’s Ratio
KRAS: Kirsten Ras Oncogene
MACROD2: Mono-ADP Ribosylhydrolase 2
mCRC: Metastatic Colorectal Cancer
mCRC-liver: CRC with Liver-Only Metastasis
mCRC-lung: CRC with Lung-Only Metastasis
mCRC-NRLN: CRC with Non-Regional Lymph Node-Only Metastasis
mCRC-peritoneum: CRC with Peritoneum-Only Metastasis
MET: MET Proto-Oncogene, Receptor Tyrosine Kinase
MLH1: DNA Mismatch Repair Protein1
MLH2: DNA Mismatch Repair Protein2
MMR: Mismatch Repair
MSH6: MutS Protein Homolog 6
MSI: Microsatellite Instability
MUC16: Mucin 16
NOTCH3: Notch Receptor 3
NRLN: Non-Regional Lymph Node
pCRC: Primary Colorectal Cancer
PIK3CA: Phosphoinositide 3 Kinase, Catalytic, Alpha Polypeptide
PLAGL2: Pleiomorphic Adenoma Gene-Like 2
PMS2: MS1 Homolog 2, Mismatch Repair System Component
RBFOX1: RNA Binding Fox-1
SYNE1: Synaptic Nuclei Expressed Gene 1
TCF7L2: Transcription Factor 7 Like 2
TCGA: The Cancer Genome Atlas
TP53: Tumor Protein P53
TSPY26P: Testis Specific Protein Y-Linked 26, Pseudogene
TTN: Titin
WWOX: WW Domain Containing Oxidoreductase
Figure S1: The survival rate of mCRC patients with only peritoneal metastasis is lower than that of patients with only liver metastasis.
Figure S2: The survival rate of mCRC patients with peritoneal only metastasis is lower than that of patients with lung only metastasis.
Figure S3: There was no statistical difference in survival rate between non-regional lymph node metastasis and peritoneal metastasis.
Figure S4: Mutation Count (log2) indicates that the number of gene mutations in patients with mCRC is less than that in patients with primary CRC.
Article Info
Article Type
Research ArticlePublication history
Received: Thu 17, Dec 2020Accepted: Sat 02, Jan 2021
Published: Fri 29, Jan 2021
Copyright
© 2023 Jaydutt V. Vadgama. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.COR.2021.01.04
Author Info
Wenhong Deng Katrina M Schrode Magda Shaheen Jaydutt V. Vadgama Yong Wu
Corresponding Author
Jaydutt V. VadgamaDivision of Cancer Research and Training, Department of Internal Medicine, Charles Drew University of Medicine and Science, David Geffen UCLA School of Medicine and UCLA Jonsson Comprehensive Cancer Center, Los Angeles, USA
Figures & Tables
Table 1: Effect of gene mutation on survival rate of CRC patients with single-site tumor metastasis (liver, lung, lymph node, and peritoneum). To reduce statistical bias, patients with gene mutation rate >=10% and cells with the number of patients >=7 were selected. “----“ means patients with gene mutation rate <10% or the number of patients <7.
|
Mutation contribute to overall survival |
Only liver metastasis |
Only lung metastasis |
Only lymph metastasis |
Only peritoneum metastasis |
|
APC mutation |
No |
Yes* |
Yes* |
No |
|
TP53 mutation |
No |
No |
No |
No |
|
KRAS mutation |
Yes* |
No |
No |
No |
|
PIK3CA mutation |
No |
No |
No |
No |
|
SMAD4 mutation |
No |
---- |
---- |
No |
|
TCF7L2 mutation |
Yes* |
---- |
No |
---- |
|
ATM mutation |
---- |
No |
---- |
---- |
|
AMER1 mutation |
---- |
No |
---- |
---- |
|
ERBB4 mutation |
---- |
---- |
---- |
No |
|
NOTCH3 mutation |
---- |
---- |
---- |
No |
|
BRAF mutation |
---- |
---- |
---- |
Yes* |
Yes* Means P<0.05 mutation type gene compared with wild type gene.
Table 2: The statistical results of (Figures 1C-1H and Figures 3A-3E).
|
|
Statistical Test |
p-Value |
|
Figure 1C Tumor Sample Histology |
Chi-squared Test |
P<0.01 |
|
Figure 1D Mutation Count |
Kruskal Wallis Test |
P<0.01 |
|
Figure 1E Age at Diagnosis |
Kruskal Wallis Test |
P<0.01 |
|
Figure 1F Primary Tumor Location |
Chi-squared Test |
P<0.01 |
|
Figure 1G MSI Status |
Chi-squared Test |
P<0.01 |
|
Figure 1H Sex |
Chi-squared Test |
P=0.969 |
|
Figure 3A MSI Status |
Chi-squared Test |
P<0.01 |
|
Figure 3B Tumor Sample Histology |
Chi-squared Test |
P<0.01 |
|
Figure 3C Primary Tumor Location |
Chi-squared Test |
P<0.01 |
|
Figure 3D Sex |
Chi-squared Test |
P=0.413 |
|
Figure 3E Age at Diagnosis |
Kruskal Wallis Test |
P<0.01 |
Table 3: Baseline characteristics of mCRC patients with liver only metastasis.
|
Variable |
Only liver metastasis(n=520) |
|
KRAS_status (%) |
|
|
Mut |
326(62.7%) |
|
Wild |
194(37.3%) |
|
TCF7L2_status (%) |
|
|
Mut |
476(91.5%) |
|
Wild |
44(8.5%) |
|
Age_at_Diagnosis,median(range),years |
53.94(24-83) |
|
Sex, n(%) |
|
|
Male |
299(57.5%) |
|
Female |
221(42.5%) |
|
Fraction_Genome_Altered |
0.21(0-0.78) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
204(39.2%) |
|
Yes |
316(60.8%) |
|
Molecular_Subtype(%) |
|
|
MSS |
507(97.5%) |
|
MSI |
11(2.1%) |
|
Unknow |
2(0.4%) |
|
MSI_Score |
1.59(0-47.7) |
|
Mutation_Count |
8.85(0-249) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
34(6.5%) |
|
Mod-Poorly-Diff |
36(6.9%) |
|
Mod_Diff |
326(62.7%) |
|
Unknow |
124(23.8%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
40(7.7%) |
|
Cecum |
51(9.8%) |
|
Descending Colon |
35(6.7%) |
|
Hepatic Flexure |
13(2.5%) |
|
Rectosigmoid |
61(11.7%) |
|
Rectum |
111(21.3%) |
|
Sigmoid Colon |
162(31.2%) |
|
Splenic Flexure |
11(2.1%) |
|
Transverse Colon |
28(5.4%) |
|
Unknow |
8(1.5%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
390(75%) |
|
Right |
124(23.8%) |
|
Unknow |
6(1.2%) |
|
Sample_Type, n(%) |
|
|
Primary |
209(40.2%) |
|
Metastasis |
311(59.8%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
32.39(-7.53-270.53) |
|
Time_to_Metastasis, median (range),month |
4.69(0-120.00) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
326(62.7%) |
|
Conventional_With_Mucinous_Component |
30(5.8%) |
|
Mucinous |
6(1.2%) |
|
PDC |
33(6.3%) |
|
Unknow |
125(24%) |
|
Overall_Survival_Months, median(range),month |
47.26(0.57-292.93) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
151(29%) |
|
Living |
369(71%) |
Table 4: COX hazard analysis of overall survival for mCRC patients with liver only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
|
P |
HR |
95%CI |
|
|||
|
down |
upper |
down |
upper |
|
||||||
|
KRAS_status |
0.014 |
1.308 |
1.056 |
1.620 |
0.863 |
0.973 |
0.716 |
1.323 |
|
|
|
TCF7L2_status |
0.951 |
0.989 |
0.705 |
1.388 |
|
0.326 |
1.239 |
0.808 |
1.901 |
|
|
Age_at_Diagnosis |
0.873 |
0.999 |
0.989 |
1.009 |
|
0.779 |
1.002 |
0.989 |
1.014 |
|
|
Sex |
0.200 |
0.872 |
0.708 |
1.075 |
|
0.037 |
1.334 |
1.018 |
1.749 |
|
|
Fraction_Genome_Altered |
0.002 |
0.348 |
0.179 |
0.676 |
|
0.343 |
0.594 |
0.203 |
1.743 |
|
|
Chemo_Exposure_of_sequenced_specimen |
≤0.001 |
0.444 |
0.358 |
0.550 |
|
0.024 |
1.454 |
1.050 |
2.015 |
|
|
Molecular_Subtype |
0.607 |
1.180 |
0.628 |
2.217 |
|
0.892 |
0.641 |
0.001 |
389.262 |
|
|
MSI_Score |
0.728 |
0.996 |
0.977 |
1.017 |
|
0.944 |
1.006 |
0.851 |
1.189 |
|
|
Mutation_Count |
0.183 |
1.003 |
0.998 |
1.008 |
|
0.875 |
1.004 |
0.950 |
1.062 |
|
|
Patient_Tumor_Grade |
0.046 |
0.802 |
0.645 |
0.997 |
|
0.426 |
0.846 |
0.560 |
1.278 |
|
|
Primary_Tumor_Site |
0.447 |
0.981 |
0.934 |
1.030 |
|
0.137 |
1.064 |
0.981 |
1.154 |
|
|
Primary_Tumor_Location |
0.041 |
1.300 |
1.011 |
1.673 |
|
0.914 |
1.025 |
0.656 |
1.601 |
|
|
Sample_Type |
≤0.001 |
0.446 |
0.357 |
0.555 |
|
0.548 |
0.903 |
0.647 |
1.260 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.822 |
0.806 |
0.838 |
|
≤0.001 |
0.811 |
0.792 |
0.831 |
|
|
Time_to_Metastasis |
0.907 |
1.000 |
0.992 |
1.007 |
|
0.732 |
1.002 |
0.992 |
1.012 |
|
|
Tumor_Sample_Histology |
0.179 |
1.111 |
0.953 |
1.294 |
|
0.891 |
1.020 |
0.768 |
1.355 |
|
Table 5: Baseline characteristics of mCRC patients with lung-only metastasis.
|
Variable
|
Only lung metastasis(n=63)
|
|
APC_status (%) |
|
|
Mut |
50(79.4%) |
|
Wild |
13(20.6%) |
|
Age_at_Diagnosis,median(range),years |
56.69(29-84) |
|
Sex, n(%) |
|
|
Male |
33(52.4%) |
|
Female |
30(47.6%) |
|
Fraction_Genome_Altered |
0.1948(0-0.57) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
24(38.1%) |
|
Yes |
39(61.9%) |
|
Molecular_Subtype(%) |
|
|
MSS |
62(98.4%) |
|
MSI |
1(1.6%) |
|
MSI_Score |
1.59(0-47.7) |
|
Mutation_Count |
7.11(1-44) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
2(3.2%) |
|
Mod-Poorly-Diff |
4(6.3%) |
|
Mod_Diff |
41(65.1%) |
|
Unknow |
16(25.4%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
1(1.6%) |
|
Cecum |
5(7.9%) |
|
Descending Colon |
3(4.8%) |
|
Rectosigmoid |
10(15.9%) |
|
Rectum |
34(54.0%) |
|
Sigmoid Colon |
9(14.3%) |
|
Transverse Colon |
1(1.6%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
57(90.5%) |
|
Right |
6(9.5%) |
|
Sample_Type, n(%) |
|
|
Primary |
26(41.3%) |
|
Metastasis |
37(58.7%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
21.25(0.90-130.67) |
|
Time_to_Metastasis, median (range),month |
18.61(0-68.20) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
38(60.3%) |
|
Conventional_With_Mucinous_Component |
5(7.9%) |
|
Mucinous |
2(3.2%) |
|
PDC |
2(3.2%) |
|
Unknow |
16(25.4%) |
|
Overall_Survival_Months, median(range),month |
36.16(1.3-138.03) |
|
Overall_Survival_Status (%) |
|
|
Deceased |
20(31.7%) |
|
Living
|
43(68.3%)
|
Table 6: COX hazard analysis of overall survival for mCRC patients with lung only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||
|
down |
upper |
|
down |
upper |
||||||
|
APC _status |
0.885 |
0.941 |
0.415 |
2.134 |
0.065 |
0.255 |
0.059 |
1.090 |
||
|
Age_at_Diagnosis |
0.420 |
1.010 |
0.985 |
1.036 |
|
0.011 |
1.055 |
1.012 |
1.099 |
|
|
Sex |
0.398 |
0.772 |
0.423 |
1.408 |
|
0.669 |
1.283 |
0.409 |
4.019 |
|
|
Fraction_Genome_Altered |
0.027 |
0.103 |
0.014 |
0.769 |
|
0.583 |
3.272 |
0.047 |
226.317 |
|
|
Chemo_Exposure_of_sequenced_specimen |
0.394 |
0.767 |
0.416 |
1.413 |
|
0.154 |
2.305 |
0.732 |
7.258 |
|
|
Molecular_Subtype |
0.945 |
1.072 |
0.146 |
7.886 |
|
0.861 |
0.080 |
0.000 |
1.598e11 |
|
|
MSI_Score |
0.608 |
0.975 |
0.884 |
1.075 |
|
0.718 |
1.186 |
0.471 |
2.984 |
|
|
Mutation_Count |
0.897 |
0.996 |
0.945 |
1.051 |
|
0.598 |
0.935 |
0.729 |
1.199 |
|
|
Patient_Tumor_Grade |
0.613 |
1.388 |
0.389 |
4.952 |
|
0.720 |
0.762 |
0.173 |
3.358 |
|
|
Primary_Tumor_Site |
0.262 |
0.898 |
0.745 |
1.083 |
|
0.021 |
0.412 |
0.194 |
0.875 |
|
|
Primary_Tumor_Location |
0.756 |
1.179 |
0.417 |
3.327 |
|
0.071 |
0.045 |
0.002 |
1.305 |
|
|
Sample_Type |
0.173 |
0.648 |
0.347 |
1.210 |
|
0.117 |
2.391 |
0.805 |
7.107 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.866 |
0.821 |
0.914 |
|
≤0.001 |
0.829 |
0.767 |
0.896 |
|
|
Time_to_Metastasis |
0.966 |
1.000 |
0.982 |
1.020 |
|
0.698 |
0.991 |
0.944 |
1.039 |
|
|
Tumor_Sample_Histology |
0.641 |
1.149 |
0.642 |
2.056 |
|
0.900 |
0.948 |
0.414 |
2.174 |
|
Table 7: Baseline characteristics of mCRC patients with lymph-only metastasis.
|
Variable |
Only lymph metastasis (n=40) |
|
APC_status (%) |
|
|
Mut |
29(72.5%) |
|
Wild |
11(27.5%) |
|
Age_at_Diagnosis,median(range),years |
51.73(26-81) |
|
Sex, n(%) |
|
|
Male |
26(65%) |
|
Female |
14(35%) |
|
Fraction_Genome_Altered |
0.198(0-0.96) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
27(67.5%) |
|
Yes |
13(32.5%) |
|
Molecular_Subtype(%) |
|
|
MSS |
35(87.5%) |
|
MSI |
4(10%) |
|
Unknow |
1(2.5%) |
|
MSI_Score |
3.70(0-37.38) |
|
Mutation_Count |
16.08(1-159) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
4(10%) |
|
Mod-Poorly-Diff |
6(15%) |
|
Mod_Diff |
18(45%) |
|
Unknow |
12(30%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
3(7.5%) |
|
Cecum |
8(20.0%) |
|
Descending Colon |
2(5%) |
|
Rectosigmoid |
4(10%) |
|
Rectum |
12(30%) |
|
Sigmoid Colon |
9(22.5%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
28(70%) |
|
Right |
12(30%) |
|
Sample_Type, n(%) |
|
|
Primary |
32(80%) |
|
Metastasis |
8(20%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
15.5(-19.63-100.8) |
|
Time_to_Metastasis, median (range),month |
14.90(0-125.8) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
19(47.5%) |
|
Conventional_With_Mucinous_Component |
4(10%) |
|
Mucinous |
1(2.5%) |
|
PDC |
3(7.5%) |
|
Signet_Ring_Cell |
1(2.5%) |
|
Unknow |
12(30%) |
|
Overall_Survival_Months, median(range),month |
28.02(0.93-126) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
16(40%) |
|
Living |
24(60%) |
Table 8: COX hazard analysis of overall survival for mCRC patients with lymph-only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||
|
down |
upper |
|
down |
upper |
||||||
|
APC _status |
0.205 |
0.462 |
0.14 |
1.523 |
0.79 |
0.665 |
0.033 |
13.407 |
||
|
Age_at_Diagnosis |
0.67 |
1.01 |
0.966 |
1.055 |
|
0.331 |
0.943 |
0.838 |
1.062 |
|
|
Sex |
0.388 |
1.488 |
0.604 |
3.665 |
|
0.7 |
1.715 |
0.11 |
26.722 |
|
|
Fraction_Genome_Altered |
0.105 |
0.171 |
0.020 |
1.446 |
|
0.982 |
0.863 |
0 |
2.68*10^5 |
|
|
Chemo_Exposure_of_sequenced_specimen |
0.023 |
0.297 |
0.104 |
0.847 |
|
0.588 |
0.472 |
0.031 |
7.113 |
|
|
Molecular_Subtype |
0.161 |
2.47 |
0.697 |
8.75 |
|
0.969 |
2.7e22 |
0 |
-- |
|
|
MSI_Score |
0.173 |
1.037 |
0.984 |
1.094 |
|
0.737 |
0.686 |
0.076 |
6.173 |
|
|
Mutation_Count |
0.592 |
0.997 |
0.986 |
1.008 |
|
0.038 |
0.565 |
0.329 |
0.969 |
|
|
Patient_Tumor_Grade |
0.367 |
0.703 |
0.326 |
1.513 |
|
0.956 |
1.055 |
0.156 |
7.14 |
|
|
Primary_Tumor_Site |
0.982 |
0.998 |
0.823 |
1.210 |
|
0.973 |
1.02 |
0.332 |
3.134 |
|
|
Primary_Tumor_Location |
0.886 |
1.068 |
0.435 |
2.618 |
|
0.546 |
5.469 |
0.022 |
1356.578 |
|
|
Sample_Type |
0.091 |
0.382 |
0.125 |
1.165 |
|
0.854 |
0 |
0 |
1.16E+36 |
|
|
Time_from_Met_Dx_to_Sequencing |
≤0.001 |
0.867 |
0.814 |
0.925 |
|
0.002 |
0.757 |
0.637 |
0.901 |
|
|
Time_to_Metastasis |
0.263 |
0.991 |
0.976 |
1.007 |
|
0.689 |
1.01 |
0.964 |
1.058 |
|
|
Tumor_Sample_Histology |
0.501 |
1.180 |
0.729 |
1.911 |
|
0.963 |
1.037 |
0.221 |
4.854 |
|
Table 9: Baseline characteristics of colorectal cancer patients with peritoneum-only metastasis.
|
Variable |
Only peritoneum metastasis(n=52) |
|
BRAF_status (%) |
|
|
Mut |
8(15.4%) |
|
Wild |
44(84.6%) |
|
Age_at_Diagnosis,median(range),years |
54.52(13-80) |
|
Sex, n(%) |
|
|
Male |
28(53.8%) |
|
Female |
24(46.2%) |
|
Fraction_Genome_Altered |
0.133(0-0.63) |
|
Chemo_Exposure_of_sequenced_specimen (%) |
|
|
No |
32(61.5%) |
|
Yes |
20(38.5%) |
|
Molecular_Subtype(%) |
|
|
MSS |
41(78.8%) |
|
MSI |
11(21.2%) |
|
MSI_Score |
6.39(0-42.22) |
|
Mutation_Count |
15.75(2-90) |
|
Patient_Tumor_Grade, n(%) |
|
|
Poorly-Diff |
9(17.3%) |
|
Mod-Poorly-Diff |
7(13.5%) |
|
Mod_Diff |
18(34.6%) |
|
Unknow |
18(34.6%) |
|
Primary_Tumor_Site, n(%) |
|
|
Ascending Colon |
6(11.5%) |
|
Cecum |
13(25%) |
|
Descending Colon |
4(7.7%) |
|
Rectosigmoid |
1(1.9%) |
|
Rectum |
5(9.6%) |
|
Sigmoid Colon |
13(25%) |
|
Hepatic Flexure |
4(7.7%) |
|
Splenic Flexure |
2(3.8%) |
|
Transverse Colon |
3(5.8%) |
|
Primary_Tumor_Location, n(%) |
|
|
Left |
26(50%) |
|
Right |
26(50%) |
|
Sample_Type, n(%) |
|
|
Primary |
30(57.7%)) |
|
Metastasis |
22(42.3%) |
|
Time_from_Met_Dx_to_Sequencing, median(range),month |
-10.59(-1365.80-159.37) |
|
Time_to_Metastasis, median (range),month |
4.49(0-62.9) |
|
Tumor_Sample_Histology(%) |
|
|
Conventional |
17(32.7%)) |
|
Conventional_With_Mucinous_Component |
7(13.5%) |
|
Mucinous |
1(1.9%) |
|
PDC |
5(9.6%) |
|
Signet_Ring_Cell |
3(5.8%) |
|
Overall_Survival_Months, median(range),month |
26.8(1.87-173.57) |
|
Overall_Survival_Status(%) |
|
|
Deceased |
24(46.2%) |
|
Living |
28(53.8%) |
Table 10: COX hazard analysis of overall survival for mCRC patients with peritoneum-only metastasis.
|
Parameter |
Univariate analysis |
|
Multivariate analysis |
|||||||||||
|
P |
HR |
95%CI |
P |
HR |
95%CI |
|||||||||
|
down |
upper |
|
down |
upper |
||||||||||
|
BRAF_status |
0.909 |
0.918 |
0.211 |
3.998 |
0.616 |
1.94 |
0.146 |
25.854 |
|
|||||
|
Age_at_Diagnosis |
0.414 |
0.989 |
0.963 |
1.016 |
|
0.001 |
0.884 |
0.821 |
0.951 |
|||||
|
Sex |
0.486 |
0.754 |
0.340 |
1.670 |
|
0.427 |
1.78 |
0.429 |
7.384 |
|||||
|
Fraction_Genome_Altered |
0.845 |
1.281 |
.108 |
15.265 |
|
0.365 |
7.989 |
0.089 |
713.333 |
|||||
|
Chemo_Exposure_of_sequenced_specimen |
0.225 |
0.597 |
0.259 |
1.374 |
|
0.015 |
22.192 |
1.815 |
271.313 |
|||||
|
Molecular_Subtype |
0.615 |
0.784 |
0.304 |
2.023 |
|
0.011 |
0 |
0 |
0.003 |
|||||
|
MSI_Score |
0.490 |
0.989 |
0.959 |
1.020 |
|
0.686 |
0.945 |
0.717 |
1.245 |
|||||
|
Mutation_Count |
0.321 |
0.992 |
0.975 |
1.008 |
|
0.004 |
1.668 |
1.177 |
2.364 |
|||||
|
Patient_Tumor_Grade |
0.192 |
0.699 |
0.408 |
1.197 |
|
0.305 |
2.408 |
0.45 |
12.885 |
|||||
|
Primary_Tumor_Site |
0.338 |
0.931 |
0.803 |
1.078 |
|
0.008 |
0.42 |
0.222 |
0.795 |
|||||
|
Primary_Tumor_Location |
0.557 |
1.257 |
.586 |
2.694 |
|
0.089 |
0.09 |
0.006 |
1.441 |
|||||
|
Sample_Type |
0.131 |
0.538 |
0.241 |
1.202 |
|
0.001 |
0.009 |
0 |
0.151 |
|||||
|
Time_from_Met_Dx_to_Sequencing |
0.081 |
0.999 |
0.998 |
1.000 |
|
0.239 |
0.997 |
0.993 |
1.002 |
|||||
|
Time_to_Metastasis |
0.593 |
1.008 |
0.978 |
1.039 |
|
0.915 |
0.994 |
0.896 |
1.103 |
|||||
|
Tumor_Sample_Histology |
0.188 |
1.227 |
0.905 |
1.663 |
|
0.303 |
1.721 |
0.613 |
4.832 |
|||||
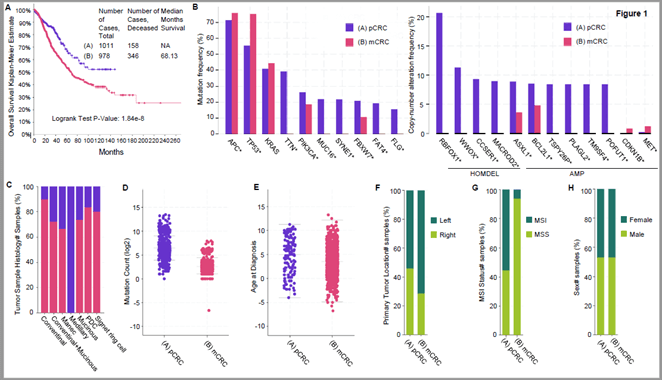
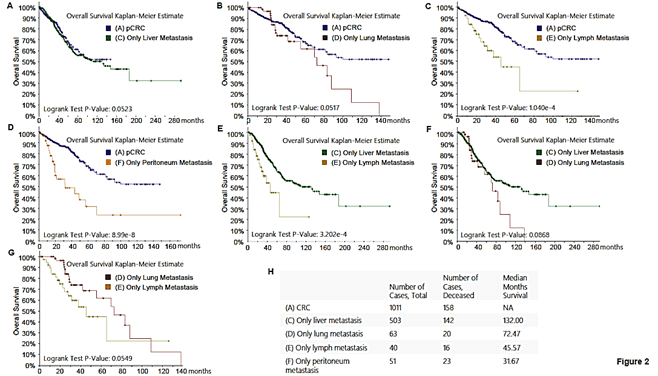
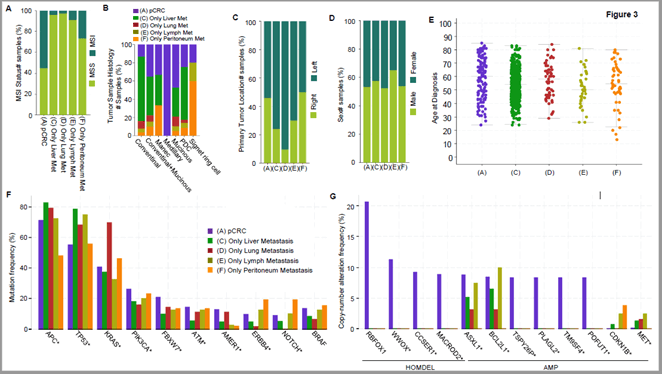
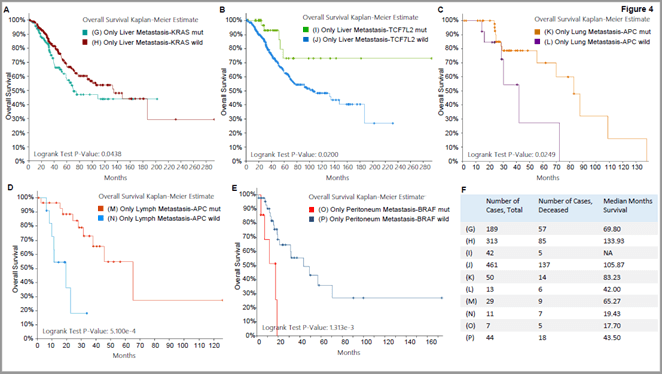
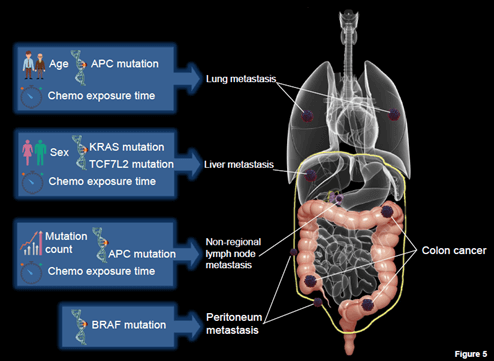
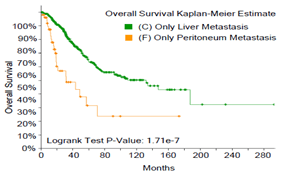
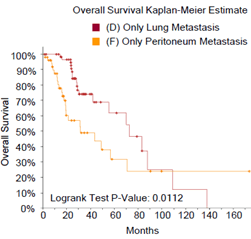
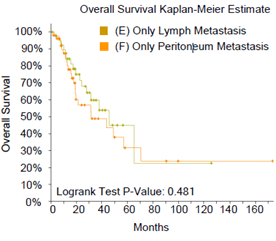
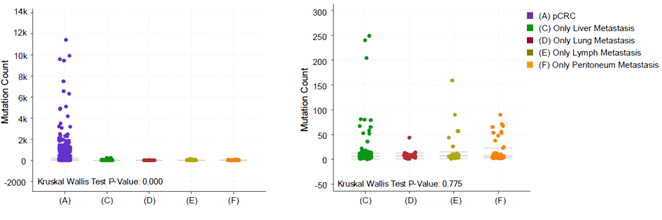
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet Tieulent J et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108. [Crossref]
- Haggar FA, Boushey RP (2009) Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 22: 191-197. [Crossref]
- Bejar L, Gili M, Diaz V, Ramirez G, Lopez J et al. (2009) Incidence and mortality by colorectal cancer in Spain during 1951-2006 and its relationship with behavioural factors. Eur J Cancer Prev 18: 436-444. [Crossref]
- Rawla P, Sunkara T, Barsouk A (2019) Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol 14: 89-103. [Crossref]
- Issa IA, Noureddine M (2017) Colorectal cancer screening: An updated review of the available options. World J Gastroenterol 23: 5086-5096. [Crossref]
- International Agency for Research on Cancer (IARC) (2012) GLOBOCAN 2012: colorectal cancer estimated incidence, mortality and prevalence worldwide in 2012. France: IARC.
- Scheithauer W, Rosen H, Kornek GV, Sebesta C, Depisch D (1993) Randomised comparison of combination chemotherapy plus supportive care with supportive care alone in patients with metastatic colorectal cancer. BMJ 306: 752-755. [Crossref]
- Eadens MJ, Grothey A (2011) Curable metastatic colorectal cancer. Curr Oncol Rep 13: 168-176. [Crossref]
- Fleming ST, Mackley HB, Camacho F, Yao N, Gusani NJ et al. (2016) Patterns of Care for Metastatic Colorectal Cancer in Appalachia, and the Clinical, Sociodemographic, and Service Provider Determinants. J Rural Health 32: 113-124. [Crossref]
- Luo Q, O'Connell DL, Kahn C, Yu XQ (2017) Colorectal cancer metastatic disease progression in Australia: A population-based analysis. Cancer Epidemiol 49: 92-100. [Crossref]
- Tsikitis VL, Malireddy K, Green EA, Christensen B, Whelan R et al. (2009) Postoperative surveillance recommendations for early stage colon cancer based on results from the clinical outcomes of surgical therapy trial. J Clin Oncol 27: 3671-3676. [Crossref]
- Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID (2014) Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol 25: 651-657. [Crossref]
- Elias D, Liberale G, Vernerey D, Pocard M, Ducreux M et al. (2005) Hepatic and extrahepatic colorectal metastases: when resectable, their localization does not matter, but their total number has a prognostic effect. Ann Surg Oncol 12: 900-909. [Crossref]
- Pulitano C, Bodingbauer M, Aldrighetti L, de Jong MC, Castillo F et al. (2011) Liver resection for colorectal metastases in presence of extrahepatic disease: results from an international multi-institutional analysis. Ann Surg Oncol 18: 1380-1388. [Crossref]
- Carpizo DR, Are C, Jarnagin W, Dematteo R, Fong Y et al. (2009) Liver resection for metastatic colorectal cancer in patients with concurrent extrahepatic disease: results in 127 patients treated at a single center. Ann Surg Oncol 16: 2138-2146. [Crossref]
- Nozue M, Oshiro Y, Kurata M, Seino K, Koike N et al. (2002) Treatment and prognosis in colorectal cancer patients with bone metastasis. Oncol Rep 9: 109-112. [Crossref]
- Koppe MJ, Boerman OC, Oyen WJ, Bleichrodt RP (2006) Peritoneal carcinomatosis of colorectal origin: incidence and current treatment strategies. Ann Surg 243: 212-222. [Crossref]
- Damiens K, Ayoub JP, Lemieux B, Aubin F, Saliba W et al. (2012) Clinical features and course of brain metastases in colorectal cancer: an experience from a single institution. Curr Oncol 19: 254-258. [Crossref]
- Riihimaki M, Hemminki A, Sundquist J, Hemminki K (2016) Patterns of metastasis in colon and rectal cancer. Sci Rep 6: 29765. [Crossref]
- Khattak MA, Martin HL, Beeke C, Price T, Carruthers S et al. (2012) Survival differences in patients with metastatic colorectal cancer and with single site metastatic disease at initial presentation: results from South Australian clinical registry for advanced colorectal cancer. Clin Colorectal Cancer 11: 247-254. [Crossref]
- Augestad KM, Bakaki PM, Rose J, Crawshaw BP, Lindsetmo RO et al. (2015) Metastatic spread pattern after curative colorectal cancer surgery. A retrospective, longitudinal analysis. Cancer Epidemiol 39: 734-744. [Crossref]
- Cho M, Akiba C, Lau C, Smith D, Telatar M et al. (2016) Impact of RAS and BRAF mutations on carcinoembryonic antigen production and pattern of colorectal metastases. World J Gastrointest Oncol 8: 128-135. [Crossref]
- Kim MJ, Lee HS, Kim JH, Kim YJ, Kwon JH et al. (2012) Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer 12: 347. [Crossref]
- Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A et al. (2018) Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 33: 125.e3-136.e3. [Crossref]
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ et al. (2018) Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 17: 291.e6-304.e6. [Crossref]
- Sanchez Vega F, Mina M, Armenia J, Chatila WK, Luna A et al. (2018) Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173: 321.e10-337.e10. [Crossref]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ et al. (2018) An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173: 400.e11-416.e11. [Crossref]
- Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G et al. (2014) Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 15: 454. [Crossref]
- Engstrand J, Nilsson H, Stromberg C, Jonas E, Freedman J (2018) Colorectal cancer liver metastases-a population-based study on incidence, management and survival. BMC Cancer 18: 78. [Crossref]
- Kawamura H, Yamaguchi T, Yano Y, Hozumi T, Takaki Y et al. (2018) Characteristics and Prognostic Factors of Bone Metastasis in Patients With Colorectal Cancer. Dis Colon Rectum 61: 673-678. [Crossref]
- Miyoshi N, Ohue M, Shingai T, Noura S, Sugimura K et al. (2015) Clinicopathological characteristics and prognosis of stage IV colorectal cancer. Mol Clin Oncol 3: 1093-1098. [Crossref]
- Hwang M, Jayakrishnan TT, Green DE, George B, Thomas JP et al. (2014) Systematic review of outcomes of patients undergoing resection for colorectal liver metastases in the setting of extra hepatic disease. Eur J Cancer 50: 1747-1757. [Crossref]
- Wang J, Li S, Liu Y, Zhang C, Li H et al. (2020) Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med 9: 361-373. [Crossref]
- Tanaka H, Deng G, Matsuzaki K, Kakar S, Kim GE et al. (2006) BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int J Cancer 118: 2765-2771. [Crossref]
- Carethers JM, Jung BH (2015) Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 149: 1177.e3-1190.e3. [Crossref]
- Minde DP, Anvarian Z, Rudiger SG, Maurice MM (2011) Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol Cancer 10: 101. [Crossref]
- Broderick P, Dobbins SE, Chubb D, Kinnersley B, Dunlop MG et al. (2017) Validation of Recently Proposed Colorectal Cancer Susceptibility Gene Variants in an Analysis of Families and Patients-a Systematic Review. Gastroenterology 152: 75.e4-77.e4. [Crossref]
- Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T et al. (2005) The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 23: 7518-7528. [Crossref]
- Yan C, Theodorescu D (2018) RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer. Pharmacol Rev 70: 1-11. [Crossref]
- Auclin E, Zaanan A, Vernerey D, Douard R, Gallois C et al. (2017) Subgroups and prognostication in stage III colon cancer: future perspectives for adjuvant therapy. Ann Oncol 28: 958-968. [Crossref]
