P28-Fur-Grb, A Novel Fusion Protein with Enhanced Targeted Cytotoxicity Against Breast Cancer
A B S T R A C T
Targeting tumor cells via multiple pathways promises the emergence of a new era in cancer therapy. Consisting of a cell-binding ligand and a cytotoxic moiety, cytolytic fusion proteins can selectively bind and kill target cells with minimal adverse effects. We designed a novel immunoproapoptotic fusion protein, p28-fur-GrB, composed of the cancer-specific azurin-derived cell penetrating peptide, p28, and a mutant version of human serine protease granzyme B. The two moieties were genetically fused by a furin sensitive linker, allowing in vivo cleavage and activation of the immunotoxin after cell entry. Synthesized coding gene of the recombinant protein was cloned and expressed in HEK293T cells, and nickel chromatography was applied for protein purification. After in vitro furin cleavage and primary analyses of SDS-PAGE, Western blotting, GrB activity and ELISA binding assay, the fusion protein was tested for its cytotoxicity on various breast cancer cell lines. Suppression of cell proliferation and viability was evaluated using the WST-1 assay. Furthermore, DNA fragmentation was measured as an indication of apoptotic effects of the fusion protein on treated cells. Based on our results, p28-fur-GrB was efficiently cleaved by furin and showed high GrB activity and binding affinity after cleavage. Following 72h of incubation with IC50 values of the fusion protein, significant cytotoxic effects of 80.6%, 77.1%, 74% and 69.6% were recorded for BT-474, MCF7, SK-BR-3 and MDA-MB-231 tumor cells, respectively. Proliferative potential of MCF 10A normal cells was not affected by the treatment. Analysis of the rate of apoptosis in treated cells confirmed our cytotoxicity results. We concluded that p28-fur-GrB is a potent anti-tumor agent with high cytotoxicity against breast cancer cells.
Keywords
Granzyme B, p28, azurin, breast cancer, fusion protein, targeting
Introduction
Despite great advancements in the fields of drug delivery and cancer therapy, most human malignancies remain unbeatable. Severe side-effects toward normal cells, systemic toxicity, drug resistance and high relapse rates have largely impaired the effectiveness of current cancer treatment strategies [1]. Targeted therapeutic approaches for the selective elimination of malignant cells have the potential to address these issues. This depends on the identification of tumor-specific ligands escorting cytotoxic moieties to tumor cells or their environment following systemic administration [2]. A novel candidate is cytolytic fusion proteins (CFPs) comprised of a ligand targeting cancer cell biomarkers fused to apoptosis-inducing effector proteins [3]. Nearly all previously developed CFPs include antibody fragments as their cell-binding moiety; although, other tumor-specific proteins have also been discovered for this purpose [4, 5].
p28, a peptide inhibitor of p53 ubiquitination derived from azurin (amino acids 50–77), a cupredoxin secreted by Pseudomonas aeruginosa, binds to and activates mutant p53, resulting in antiproliferative activity [6]. Targeting overexpressed p53 in malignant cells has no effect on wild-type p53 in normal cells because wild-type p53 is properly folded and expressed at low levels ensured by HDM2. Binding of p28 to a hydrophobic, non-mutable region within the DNA-binding domain of p53, inhibits proteasomal degradation via an HDM2-independent pathway, inducing a post-translational increase in p53 and elevating the cyclin-dependent kinase inhibitors p21 and p27, which in turn reduces the level of CDK2 and cyclin A1, leading to G2–M cell-cycle arrest and subsequent programmed cell death [7, 8]. This anionic cell-penetrating peptide contains an amphipathic α-helical motif that allows it to preferentially enter tumor cells and confer selective oncolytic activity [9]. Suggested entry routes for p28 include aberrant N-glycosylated integrins and cadherins on the cancer cell surface, an energy-dependent endocytotic or pore-related process, binding to lipid micro domain (raft) components, localization in caveolae and eventual moving to the Golgi, endoplasmic reticulum, and nucleus [10]. Extensive kinetic analysis indicates that following plasma membrane penetration, p28 reaches late endosomes, lysosomes, and the Golgi associated with caveolae in a dynamin-independent clathrin-independent carrier-mediated manner [11]. Two phase I clinical trials against adult stage IV solid tumors and pediatric CNS tumors, proved high efficacy, safety and tolerability of this peptide [8, 12].
The effector component of a CFP may include an RNase, kinase, protease or another chemotherapeutic agent. Granzyme B (GrB), a 32-KD human serine protease involved in the granule-mediated apoptosis of virus-infected or transformed tumor cells by effector cells of the innate and adaptive immune system, has several inherent advantages as an immunotoxin, including high cytotoxic efficacy, a broad portfolio of apoptosis inducing mechanisms and low immunogenicity [13]. Yet, natural inhibition by serpin B9, a high isoelectric point resulting in a positive surface charge contributing to non-specific binding to normal cells, and relatively slow release after uptake by the receptor-mediated endocytosis have limited the clinical application of GrB [14]. Cytotoxic activity of GrB generally involves the induction of several mechanisms that interfere with carcinogenesis and tumor cell proliferation. Cleaving and direct activation of several procaspases and downstream caspase substrates such as the inhibitor of caspase-activated DNase, direct damage to the mitochondrial structure and cleaving poly (ADP-ribose) polymerase and nuclear matrix antigen by rapid translocation to the nucleus, are the main mechanisms of action proposed for GrB. Furthermore, as a potent proapoptotic agent, GrB can positively affect radio-sensitivity, metastatic spread and sensitivity to chemotherapy toward improved clinical outcome [2, 15].
Herein, we introduce the immunoproapoptotic fusion protein, p28-fur-GrB, as a novel anticancer agent with selective cytotoxicity against breast cancer cells. Previously, we designed a chimeric protein composed of GrB and azurin [16]. Our results showed selective cytotoxicity of the construct in breast cancer cells. Although, nonspecific binding to non-target cells and molecules, insensitivity of some cancer cells to GrB, and the immunogenicity associated with azurin, encouraged us to upgrade the former design to another fusion protein, in which azurin is replaced with p28. Connected to a genetically enhanced mutant of GrB via a furin recognition motif, p28 was used to specifically target cancer cells. As a cellular endoprotease, furin localizes principally to the trans-Golgi network, the cell surface, and early endosomes, proteolytically activating large numbers of secreted proteins [17]. We predicted that following selective cell entry via p28 and furin cleavage in endosome, the recombinant GrB and p28 could translocate into the cytosol, leading to irreversible cell death. This hypothesis was tested using bio-physico-chemical analyses, leading to promising results.
Materials & Methods
I Construction of expression plasmid
A mutant GrB gene was designed, in which coding sequence of the residue R201 was replaced with a lysine codon, and the residues R96, R100, R102, K221, K222, K225 and R226 were exchanged for an alanine. The codon adaptation index for the p28, R9 and GrB genes was calculated as a measure for the potential influence of different codon usage on the expression of p28-fur-GrB fusion protein in a mammalian host using a free online tool accessible at www.genscript.com/cgi-bin/tools/rare_codon_analysis (GenScript, Piscataway, NJ). Synthesized genes encoding p28, R9 (furin cleavage site) and GrB were cloned downstream and in frame with DNA encoding a signal sequence in the expression vector. Vector sequences were confirmed by DNA sequencing.
II Cell lines
Human embryonic kidney 293 cells (HEK293T, ATCC® HTB-30™) were cultivated in Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich, USA) supplemented with 15% fetal bovine serum (FBS, Sigma-Aldrich, USA), 2.5µg/ml insulin, 100 unit/mL penicillin (Sigma-Aldrich, USA), and 100 µg/mL streptomycin (Sigma-Aldrich, USA). MCF 10A (ATCC® CRL-10317), MDA-MB-231 (ATCC® HTB-26), MCF7 (ATCC® HTB-22) and SKBR3 (ATCC® HTB-30) were grown according to ATTC culture method. The medium was McCoy's 5A Modified Medium (Thermo Fisher Scientific, USA), supplemented with 10% FBS (Sigma-Aldrich, USA). All cells were cultured on optimum conditions of 37ºC and 5% CO2. Prior to passage, cells were washed three time with PBS (Gibco, USA), and then were detached by 0.25% trypsin (Sigma-Aldrich, USA) for 5min at RT. After centrifugation at 300g for 5min, cells were resuspended in fresh DMEM and cultured.
III HEK293T Transfection
In order to prepare a stable clone of HEK293T expressing p28-fur-GrB fusion protein, we used the PiggyBac system. Two plasmids, PB-Cuo-MCS-IRES-GFP-EF1α-CymR-Puro (System Biosciences, USA) harboring the recombinant protein and PiggyBac transposase vector (System Biosciences, USA), were transfected by lipofectamine 3000 (Thermo Fisher Scientific, USA) into HEK293T cells in a 6-well plate. Finally, positive clones were selected by 1.1 μg/mL puromycin (InvivoGen, USA) and used for protein purification.
IV Protein Expression and Purification
Around 1 × 106 cells of transfected HEK293T were seeded in ten T150 flasks and cultivated as described above. A strong signal sequence at the N-terminal of the fusion protein caused cells to secrete recombinant protein into media. Every 24h, media were changed, and removed media were centrifuged at 300 g to remove floating cells, and dialyzed against phosphate-buffered saline (PBS) supplemented with 10 mM imidazole. After 48h of dialysis, media were collected and concentrated by 3-KDa Amicon Ultra Centrifugal Filter Units. Protein purification was carried out by immobilized-metal affinity chromatography (IMAC) using a Ni-NTA superflow column (Qiagen, Germany). Concentrated media were passed through pre-equilibrated Ni2+ column. To remove unbound proteins, column was washed three time with increasing concentration of imidazole, 20mM, 40mM and 60mM. Finally, recombinant p28-fur-GrB were eluted by 300mM imidazole.
V Furin cleavage assay
Furin cleavage assay was performed as previously described [17]. Concentrated cell culture supernatants from the previous step were centrifuged at 3000 g for 30 min at 4 ºC and through Amicon Ultra concentrators (30000 MWCO, Millipore, USA), to exchange buffers to what were applied in the later furin cleavage assay. All the reaction buffer solutions contained final concentrations of 3 mmol/L CaC12 and 0.1% Triton X-100. Respectively, 50 mmol/L sodium acetate buffer and 10 mmol/L HEPES buffer were added for pH 5.4 and pH 7.2. The final total reaction mixtures were 100 µL, each containing 200 ng of recombinant human furin (ab167741, Abcam, UK). Reaction mixtures were incubated for 16 h at room temperature. Subsequently, SDS loading buffer was added to quench the reaction, and proteins were analyzed by SDS-PAGE and autoradiography. The percentage of furin cleavage rate was calculated using Glyko Bandscan software (Version 5.0) from the following formula:
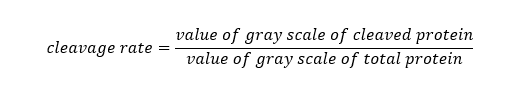
VI Anti-p28 Antibody
By introducing a cysteine at the NH2 terminus, p28 peptide was conjugated with keyhole limpet hemocyanin through the thiol groups of the cysteine residue. Subsequently, polyclonal antibody against p28 was generated in rabbits (New Zealand White). Antibody titer was determined by direct ELISA using 0-3 µg per well p28. An antibody dilution of 1:150,000 was sufficient to give a reproducible change in absorbance of 0.5 at 450 nm after 15 min incubation with substrate (1-Step PNPP; Pierce) at 25°C when 96-well plates (Nunc) were coated with 1 µg per well p28.
VII SDS-PAGE and Western Blotting
The purified p28-fur-GrB was analyzed by 12% denaturing polyacrylamide gel electrophoresis (SDS-PAGE), as previously described [18]. Protein bands were stained using Coomassie Brilliant Blue G-250 (Thermo Fisher Scientific, USA). Resolved proteins were transferred onto pre-activated polyvinylidene difluoride (PVDF) membrane via wet transfer system using Towbin buffer (25 mM Tris, 192 mM glycine, 20% (v/v) methanol, pH 8.3) at 4 ºC. After 2.5h, membrane was blocked by blocking buffer (7.5% skim milk, 0.1% tween 20, PBS) for 1h at room temperatures. Cleaved and uncleaved fusion proteins were detected using a diluted mixture of rabbit anti-6X His tag (ab137839, Abcam, UK) and anti-p28 antibody as primary antibodies and incubated at 4 ºC overnight. Finally, goat anti-rabbit HRP-conjugated secondary antibody (ab6721, Abcam, UK), was added. After 1h, 0.06% 3,3'-Diaminobenzidine (DAB) in PBS solution was added. Subsequent to each antibody incubation, membrane was washed 6 times with PBS, 5 min per wash.
VIII GrB Activity Assay
Activated GrB from the furin cleavage step was analyzed for its enzymatic activity. The following procedures were performed on ice unless stated otherwise. Samples were triplicated in 96-well plates. A stock solution of Ac-IETD-pNA (Calbiochem, Merck, USA) in Me2SO, a synthetic peptide substrate of GrB, was diluted with the buffer at a final concentration of 0.2 mM (0.2 M HEPES, 0.3 M NaCl, 1 mM EDTA, 0.5% (v/v) Triton X-100, pH 7.0). After incubation for 5 min at 27°C, substrate hydrolysis was measured by colorimetric absorbance changes at 405 nm.
IX Enzyme-linked immunosorbent assay (ELISA)
For evaluating the binding properties of GrB and p28 proteins, 50 ng of cleaved fusion protein, BSA (Sigma-Aldrich, USA) and HER2 (Merck, USA) were coated onto 96-well Maxisorp immunoplates at 4ºC. After 16h, the plates were washed once to remove unbound protein. Then, wells were blocked by adding 300 µL of blocking buffer (5% (w/v) skim milk-PBS) and incubated at room temperature for 1h on an orbital shaker. After discarding blocking buffer, 100 µL of primary antibody, anti-granzyme B antibody (ab4059, Abcam, UK), and secondary antibody, HRP conjugated goat anti-rabbit IgG (ab6721, Abcam, UK), were sequentially added to detect GrB protein. After each step, wells were washed 5 times to remove unbound antibodies. In final step, 50 µL of TMB substrate solution (Thermo Fisher Scientific, USA) was added to each well and incubated at room temperature in darkness for 20-30 min. The HRP reaction was terminated by adding 50 µL of 1 M HCl. Alternatively, rabbit anti-p28 antibody and HRP conjugated goat anti-rabbit IgG (ab6721, Abcam, UK) were used for the detection of p28, with other steps followed similar to the aforementioned method.
X WST-1 Assay
The IC50 of p28-fur-GrB recombinant protein was measured. Around 7000 cells were seeded into a 96-well plate and incubated for 24h prior to treatment. Next, increasing concentrations of the fusion protein, 1 nM, 10 nM, 20 nM, 50 nM, 100 nM, 200 nM, 500 nM were added to each well and incubation continued for 72h. To count the viable cells in each well, 10 µL of WST-1 (water-soluble tetrazolium) was added to each well. After 2h, absorbance was measured at 440 nm, as recommended by the manufacturer (Sigma-Aldrich, USA). Cells treated with 1% Triton X-100 were regarded as positive control.
XI DNA Fragmentation Assay
The rate of apoptosis in treated cells was analyzed using the DNA fragmentation assay, as described [19]. Briefly, tumor cells were labeled with 1 µCi of [3H] thymidine overnight. Cells were washed three times, pre-treated with 1 mg/ml of the apoptotic stimulus p28-fur-GrB fusion protein and then incubated for 2-16 hr at 37 °C. Assays were performed with 1000 cells per well and in triplicate. The amount of [3H]thymidine released by the target cells represents the amount of fragmented DNA and is calculated by using the following formula:
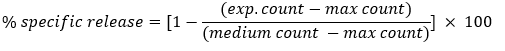
The max count signifies the amount of label retained after incubation for 6h with 1M HCl. Unlabeled thymidine (1.5 mM) was added to ensure that the released thymidine is not incorporated into other nuclei.
XII Statistics
Data are expressed as mean values and standard deviation of at least three independent experiments. Student’s two-tailed t-tests were used to determine the statistical significance of the data using GraphPad Prism v4.0 and a significance threshold of p < 0.05.
Results & Discussion
Despite the significant decline in cancer mortality over the past two decades, breast cancer is still the most common form of malignancy in woman, accounting for about 30% of all new cancer diagnoses [20]. Current treatment options include radical surgery, radiotherapy and chemotherapy. Besides severe side effects on normal tissues and organs, cytotoxic chemotherapy is usually accompanied by a relapse that takes place due to the ongoing growth of tumor cells that are resistant to the chemical agent [21]. While, several targeted therapies have been developed against breast cancer, the emergence of drug resistance and systemic toxicity associated with immunotoxins and monoclonal antibodies, impairs their clinical effectiveness [22, 23].
In a previous study, we evaluated the potential of GrB-azurin as a selective eradicator of cancer cells [16]. Given the positive yet improvable results obtained following bio-physico-chemical analyses, we decided to consider some adjustments in our original design. Thus, azurin was substituted with a smaller, less immunogenic subsidiary peptide, p28, which is well-known for its selective entry to cancer cells and induction of proapoptotic effects via interaction with tumor protein p53. Since more than 50% of human breast cancers are positive for wild-type or mutant p53, oncotoxic agents such as p28, capable of targeting breast cancer cells based on their reliability on p53, are highly valued [24].
We fused p28 to a genetically enhanced mutant version of the serine protease granzyme B. GrB is one the most potent effector molecules with minimized unwanted immune responses due to the human origin of the enzyme. However, the efficacy of GrB is diminished by the endogenous inhibitor PI-9, expressed by some tumor cell lines as escape mechanism [25]. We addressed this issue by exchanging arginine residue at position 201 (R201) for a lysine (R201K), as previously described by Losasso et al [26]. Furthermore, by introducing multiple mutations in the primary amino acid sequence of GrB (R96A, R100A, R102A, K221A, K222A, K225A and R226A), we aimed at reducing the surface charge and subsequent non-specific binding of the fusion protein; a justified and well-established approach towards improving clinical competence of therapeutic proteins [27]. Figure 1 displays schematics of the expression vector and amino acid sequence of the chimeric protein.
Figure 1: Schematics of the expression construct and p28-fur-GrB fusion protein. A) Amino acid sequence of the fusion protein. The arginine residue at position 201 (R201) of GrB was exchanged for a lysine (R201K) to elicit PI-9 resistance. Seven other mutations (R96A, R100A, R102A, K221A, K222A, K225A and R226A) were introduced to the amino acid sequence of GrB to reduce surface charge of the molecule. The strong Ig kappa leader sequence was placed at the N-terminal to allow secretion of the recombinant protein to the media, and for protein purification, a C-terminal His-tag was added. B) The coding gene of p28-fur-GrB was cloned into the multiple cloning site (MCS) of PB-Cuo-MCS-IRES-GFP-EF1α-CymR-Puro expression vector.
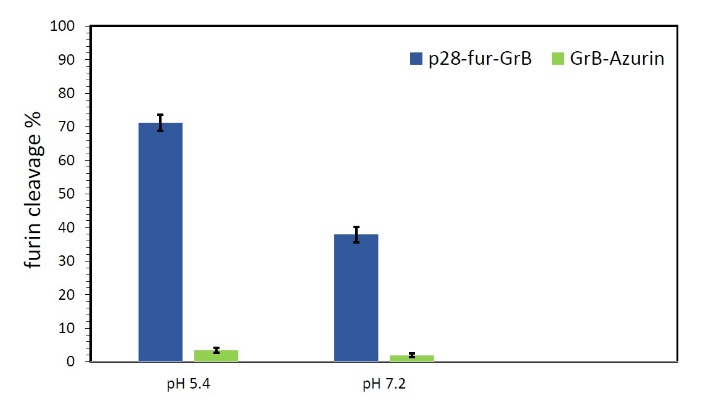
The chimeric protein was expressed in HEK293T cells and purified using affinity chromatography. Furin cleavage assay was performed to assess the sensitivity of the furin-sensitive linker and to obtain an in vitro evaluation of the cleavage process which was expected to happen in vivo. Our results corroborated with those reported by Wang et al. with successful cleavage rates of 37.9% and 71.2% at pH 7.2 and 5.4, respectively [17]. As expected, the negative control GrB-Azurin recombinant protein, which lacks a furin-sensitive site, was not cleaved at either pH (Figure 2). Thus, we concluded that p28-fur-GrB is capable of being efficiently cleaved by intracellular furins at acidic or neutral environment of endosome or cytoplasm and release the two components of the chimeric protein.
Figure 2: The results of furin cleavage assay on p28-fur-GrB, using recombinant furin at pH 7.2 and 5.4. GrB-Azurin was used as negative control. All tests were performed in triplicate.
Mainly localized to the trans-Golgi network, the cell surface and early endosomes, furin is a cellular endoprotease which has been associated with proteolytically activating many secreted proteins. Furin-induced cleavable linkers have been applied in various immunoproapoptotic fusion proteins, resulting in enhanced cytotoxicity [28, 29]. Wang et al. designed a synthetic polyarginine tract for furin cleavage between an scFv and granzyme B. The fusion protein was effectively cleaved at the furin cleavage site within the endosome, translocated to cytosol to provoke cell death, and reduced tumor size in nude mice [17]. An optimal linker can provide many advantages for improved bioactivity and specificity of the fusion protein, potentially facilitating its activity by avoiding steric hindrance or conformational changes [13]. The furin-sensitive polyarginine linker we used to connect p28 to GrB, enabled in vivo activation of the fusion protein in tumor cells, which may lead to reduced off-target binding and systemic toxicity of the drug protein [30]. Furthermore, higher cleavage rate of the polyarginine linker at a lower pH (5.4), can be interpreted as a positive point, given the low pH range at tumor microenvironment. There were preliminary concerns over possible diminishing effects of the residues remained after proteolytic cleavage at the N-terminal of GrB; although, our data demonstrates efficient bioactivity of GrB upon cleavage both in vitro and in vivo.
The proteolytic activity of granzyme B is an essential feature of the construct. It functions by initiating a cascade of apoptotic interactions, including the proteolytic cleavage of different substrates and the activation of procaspases 3 and procaspases 7. The caspase cascade continues by the processing of downstream substrates such as poly(ADP-ribose) polymerase, DNA-dependent protein kinase and the inhibitor of caspase-activated DNase [2]. In parallel, GrB cleaves caspase-independent proteins such as the mitochondrial BH3-only protein (Bid), leading to the activation of the pro-apoptotic Bcl-2 family proteins Bax/Bax, which in turn increases the permeability of the mitochondrial outer membrane and triggers the release of cytochrome C [31, 32]. GrB also cleaves parts of the cytoskeleton, such as lamin B, PARP and the C-terminus of α-tubulin [15].
Mainly localized to the trans-Golgi network, the cell surface and early endosomes, furin is a cellular endoprotease which has been associated with proteolytically activating many secreted proteins. Furin-induced cleavable linkers have been applied in various immunoproapoptotic fusion proteins, resulting in enhanced cytotoxicity [28, 29]. Wang et al. designed a synthetic polyarginine tract for furin cleavage between an scFv and granzyme B. The fusion protein was effectively cleaved at the furin cleavage site within the endosome, translocated to cytosol to provoke cell death, and reduced tumor size in nude mice [17]. An optimal linker can provide many advantages for improved bioactivity and specificity of the fusion protein, potentially facilitating its activity by avoiding steric hindrance or conformational changes [13]. The furin-sensitive polyarginine linker we used to connect p28 to GrB, enabled in vivo activation of the fusion protein in tumor cells, which may lead to reduced off-target binding and systemic toxicity of the drug protein [30]. Furthermore, higher cleavage rate of the polyarginine linker at a lower pH (5.4), can be interpreted as a positive point, given the low pH range at tumor microenvironment. There were preliminary concerns over possible diminishing effects of the residues remained after proteolytic cleavage at the N-terminal of GrB; although, our data demonstrates efficient bioactivity of GrB upon cleavage both in vitro and in vivo.
We analyzed the cleaved fusion protein for its proteolytic activity using the synthetic substrate Ac-IETD-pNA, which mimics procaspase-3 in a colorimetric substrate assay. The results implied expression of the fully active mutant GrB and retaining of its enzymatic activity following furin cleavage. As depicted in (Figure 3), we observed that the mutant version of the enzyme exhibited proteolytic activity comparable to that previously reported for the wild-type GrB, and similar to that recorded for an equimolar amount of commercial granzyme B standard.
Figure 3: The enzymatic activity of p28-fur-GrB. The granzyme B-specific substrate Ac-IETD-pNA was incubated with the uncleaved p28-fur-GrB (triangles), the cleaved p28-fur-GrB (dots) and the granzyme B standard (rectangles). The enzymatic activity was determined using a colorimetric assay and the absorbance at 405 nm was monitored for 60 min in 5-min intervals.
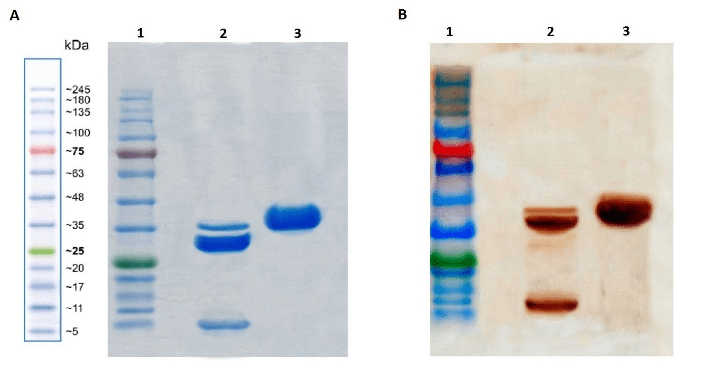
Next, purified intact p28-fur-GrB fusion protein, along with the furin-cleaved sample were analyzed by SDS-PAGE and Western blotting, as described above. Approximate molecular weights of 36, 32, and 3 KD were estimated for the expressed chimeric protein, GrB and p28, respectively (Figure 4). We achieved high recombinant protein yields of 12-16 mg/L, with a purity of 81-83% (data not shown).
Figure 4: SDS-PAGE and Western blotting analysis of the fusion protein before and after furin cleavage. (A) 12% polyacrylamide gel electrophoresis; 1-Marker; 2-p28-fur-GrB after furin cleavage; 3-uncleaved p28-fur-GrB. (B) Western blotting against p28 and his-tagged GrB; 1-Marker; 2-p28-fur-GrB after furin cleavage; 3-uncleaved p28-fur-GrB. The protein band detected at 32 KDa on lane 2 belongs to the uncleaved fusion protein after furin treatment.
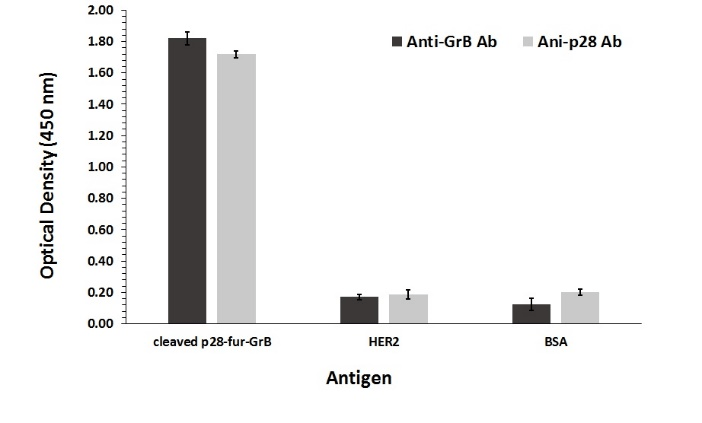
ELISA binding assay was carried out as a quantitative evaluation of the binding capability of the fusion protein components. The results showed binding of GrB and p28 to their respective ligands with relatively strong binding affinities (Figure 5).
Figure 5: ELISA binding assay. Anti-GrB and anti-p28 antibodies were applied to analyze binding properties of 50µL of the p28-fur-GrB fusion protein. HER2 and BSA were used as negative control. Measurements were performed in triplicate and error bars represent SD.
Cytotoxicity of the fusion protein in terms of suppression of cell proliferation and viability was evaluated by monitoring the oxidoreductase enzyme activity. WST-1 is a tetrazolium-derived salt that is broken to formazan within viable cells, providing a reliable and sensitive method for cell viability assessment. Following treatment of breast cancer cell lines with various concentrations of p28-fur-GrB, IC50 values of 49.7, 63.8, 85.2 and 114.1 nM were documented for BT-474, MCF7, SK-BR-3 and MDA-MB-231 cell lines, respectively. MCF 10A cells showed insignificant decrease in viability and proliferation after treatment with the chimeric protein. Considerably lower IC50 values recoded for the breast cancer cells lines compared to those previously reported for GrB-Azurin, highlighted a significant improvement in inhibitory effect and potency of the construct as a result of the abovementioned adjustments [16].
For the cell viability assay, pre-cultured cells were incubated with IC50 values of p28-fur-GrB for 72h. In parallel, cells were also treated with p28, GrB or GrB-Azurin, to account for the synergistic effect. Viable cells were counted and we observed that treatment with p28-fur-GrB led to significant cytotoxicity in BT-474 (80.6 ± 2.4%), MCF7 (77.1 ± 5.2%), SK-BR-3 (74 ± 4.5%) and MDA-MB-231 (69.6 ± 5.2%) cells compared to the untreated control, while proliferative potential of MCF 10A cells was not affected at this concentration (Figure 6). Synergistic effects of the two killer moieties (p28 and GrB) significantly improved cytotoxicity of the treatment regimen in all breast cancer cell lines, in comparison to p28, GrB or GrB-Azurin. Expectedly, p28 alone inflicted considerable cytotoxic effects on all tested cells, except for the p53-negative MDA-MB-231 (32.7 ± 2.4%) and MCF 10A (8.1 ± 1.3%) normal cells. Cells treated with GrB showed low cytotoxicity, possibly due to the lack of a transporter partner to pass GrB through the cell membrane. GrB-Azurin was moderately effective on all breast cancer cell models, yet our modifications of the construct seem to have noticeably improved cytotoxic potential of p28-fur-GrB against cancer.
Figure 6: Cell viability assay. (A) IC50 values were measured for each cell line following 72h of treatment with various concentrations of p28-fur-GrB. Proliferative potential of all breast tumor cells was dramatically inhibited, while MCF 10A normal cells were insignificantly affected by the fusion protein. (B) Medium of log-phase cells was replaced with medium containing IC50 values of p28-fur-GrB, GrB-Azurin, GrB or p28. After 72 h, the inhibitory effect of the treatment on the growth of cells in culture was determined using WST-1 cytotoxicity assay. The antiproliferative effect of p28-fur-GrB on the tested cells was dose- and time-dependent (P < 0.05). Pre-set concentrations of 50 µM, 100 nM and 500 nM were applied for p28, GrB and GrB-Azurin treatments, respectively. Cells treated with 1% Triton X-100 were used as internal control. Cell number in untreated (control) wells was considered as 100%.
Induction of apoptotic death in cells treated with p28-fur-GrB or GrB-Azurin fusion protein was evaluated using the DNA fragmentation assay. According to the data presented in (Figure 7), following treatment with p28-fur-GrB, all tested breast cancer cell lines exhibited significant increase in DNA fragmentation at 16h post treatment, while MCF 10A normal breast cells showed no apoptotic effects (p ˂ 0.05). Consistent with our cytotoxicity results, BT-474 (86.8%), MCF7 (80.9%), SK-BR-3 (74%), and MDA-MB-231 cells (71.1%) proved more sensitive to p28-fur-GrB, respectively. Both fusion proteins showed minimal cytotoxic effects on normal cells, yet after 16h of treatment, all four tumor cells demonstrated significantly improved apoptotic effects toward p28-fur-GrB, in comparison to GrB-Azurin
Figure 7: DNA fragmentation assay. Apoptotic effect of p28-fur-GrB was analyzed by measuring the amount of [3H]thymidine released by the target cells. After 2-16h of incubation with the fusion protein, all breast cancer cell lines exhibited signs of apoptotic cell death. The rate of apoptosis in treated cells was time-dependent. Compared to GrB-Azurin, our novel construct (p28-fur-GrB) showed significantly improved cytotoxic effects. Untreated cells were used as negative control.
Previously, several cytolytic fusion proteins with GrB as their cytotoxic moiety have been developed. In one study, GrB was genetically fused to an antibody fragment via a polyionic adapter peptide, requiring Cathepsin C-mediated in vivo cleavage. They aimed at targeting Lewis Y positive SK-BR-3 and MCF7 cells, which resulted in significant cytotoxicity in both cell lines with IC50 values of 35-140 nM [33]. Wang et al. produced an immunoproapoptotic protein composed of an anti-HER2 single chain antibody (e23Fv) and GrB, linked by a furin-sensitive polyarginine sequence (R9). The construct was tested against HER2-overexpressing breast cancer cells, and the results indicated efficient in vivo cleavage and translocation of the chimeric protein to the cytosol of tumor cells, leading to selective killing of tumor cells [17]. Amoury et al. fused a PI-9 resistant GrB mutant to an EpCAM-selective single-chain antibody fragment and effectively induced apoptosis in triple-negative breast cancer cells [3].
Preferential entry and induction of cell death in human breast cancer cell lines by p28, with reported IC50 values of 50-100 µM, have been described [6, 10, 24]. The cell penetrating peptide was evaluated in a clinical trial for p53 (+) metastatic solid tumors and recurrent or progressive central nervous system tumors [8, 12]. Our results demonstrated enhanced cytotoxicity and apoptotic induction by a p28-containing construct. Besides playing the vital role of a selective entry key to tumor cells, p28 assisted in induction of apoptotic response in these cells to ensure overcoming of a potential resistance. Given the heterogeneous nature of tumor microenvironment, targeting cancer from different angles, similar to how p28-fur-GrB functions, sounds like a logical approach. However, further questions concerning the safety and clinical efficacy of the designed fusion protein ought to be addressed.
A 40% drop in death rates for breast cancer since 1989, mostly due to advances in early detection and treatment, highlights the importance and potential impact of further pursuit for novel therapeutic platforms against this malignancy [20]. The golden standard in oncological therapy is exclusive targeting of abnormal cells, while leaving normal tissues and organs intact [23]. Our data implies a significant improvement in effectivity and a palpable reduction in off-target binding of the designed construct, instigated by replacing azurin with p28, using an enhanced GrB mutant and linking the two anti-tumor molecules with an in vivo cleavable furin sequence instead of the former (G4S)4 linker. Based on our findings, p28-fur-GrB is an excellent candidate for the treatment of breast cancer, with minimal unwanted side-effects. Consistent cytotoxicity results even in triple-negative breast cancer cells, promises applicability of the fusion protein against a wide range of cancers. Further preclinical research and in vivo studies are being designed to ensure safety and efficiency of this fusion protein for clinical applications. In our fight against cancer, testing novel fusion proteins in vitro can play a vital role. Although containing much more information about the effect of a potential biodrug, comprehensive in vivo analyses entail fairly large investments, both of time and finance. We believe that by facilitating the process of examination of biotherapeutics, and by investing more on the preliminary stages of in vitro bio-physico-chemical examination, many unnecessary expenses could be eliminated, thus improving our chance to find more efficient anti-cancer therapeutics.
Acknowledgment
The authors declare that no conflict of interest exists regarding the publication of this article.
Article Info
Article Type
Research ArticlePublication history
Received: Sat 07, Sep 2019Accepted: Thu 10, Oct 2019
Published: Mon 28, Oct 2019
Copyright
© 2023 Saeed Ranjbar. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.COR.2019.05.02
Author Info
Aria Momeni Azadeh Reshadmanesh Azita Fakhravar Nafiseh Paydarnia Saeed Ranjbar
Corresponding Author
Saeed RanjbarDepartment of Medical Biotechnology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
Figures & Tables




References
- Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E (2018) The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis 35: 309-318. [Crossref]
- Rosenblum M, Barth S (2009) Development of novel, highly cytotoxic fusion constructs containing granzyme B: unique mechanisms and functions. Curr Pharm Des 15: 2676-2692. [Crossref]
- Amoury M, Kolberg K, Pham AT, Hristodorov D, Mladenov R et al. (2016) Granzyme B-based cytolytic fusion protein targeting EpCAM specifically kills triple negative breast cancer cells in vitro and inhibits tumor growth in a subcutaneous mouse tumor model. Cancer Lett 372: 201-209. [Crossref]
- Hlongwane P, Mungra N, Madheswaran S, Akinrinmade O, Chetty S et al. (2018) Human granzyme B based targeted cytolytic fusion proteins. Biomedicines 6. [Crossref]
- Mungra N, Jordaan S, Hlongwane P, Naran K, Chetty S et al. (2019) Targeted human cytolytic fusion proteins at the cutting edge: harnessing the apoptosis-inducing properties of human enzymes for the selective elimination of tumor cells. Oncotarget 10: 897-915. [Crossref]
- Yamada T, Mehta RR, Lekmine F, Christov K, King ML et al. (2009) A peptide fragment of azurin induces a p53-mediated cell cycle arrest in human breast cancer cells. Mol Cancer Ther 8: 2947-2958. [Crossref]
- Bernardes N, Ribeiro AS, Seruca R, Paredes J, Fialho AM (2011) Bacterial protein azurin as a new candidate drug to treat untreatable breast cancers. Portuguese Biomed Engg Meeting 2011: IEEE.
- Warso M, Richards J, Mehta D, Christov K, Schaeffer C et al. (2013) A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br J Cancer 108: 1061-1070. [Crossref]
- Bernardes N, Seruca R, Chakrabarty AM, Fialho AM (2010) Microbial-based therapy of cancer: current progress and future prospects. Bioengineered Bugs 1: 178-190. [Crossref]
- Taylor BN, Mehta RR, Yamada T, Lekmine F, Christov K et al. (2009) Noncationic peptides obtained from azurin preferentially enter cancer cells. Cancer Res 69: 537-546. [Crossref]
- Kurrikoff K, Aphkhazava D, Langel Ü (2019) The future of peptides in cancer treatment. Curr Opin Pharmacol 47: 27-32. [Crossref]
- Lulla RR, Goldman S, Yamada T, Beattie CW, Bressler L et al. (2016) Phase I trial of p28 (NSC745104), a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in pediatric patients with recurrent or progressive central nervous system tumors: A Pediatric Brain Tumor Consortium Study. Neuro Oncol 18: 1319-1325. [Crossref]
- Hehmann-Titt G, Schiffer S, Berges N, Melmer G, Barth S (2013) Improving the therapeutic potential of human granzyme B for targeted cancer therapy. Antibodies 2: 19-49.
- Kurschus FC, Jenne DE (2010) Delivery and therapeutic potential of human granzyme B. Immunol Rev 235: 159-171. [Crossref]
- Lord SJ, Rajotte RV, Korbutt GS, Bleackley RC (2003) Granzyme B: a natural born killer. Immunol Rev 193: 31-38. [Crossref]
- Paydarnia N, Nikkhoi SK, Fakhravar A, Mehdiabdol M, Heydarzadeh H et al. (2019) Synergistic effect of granzyme B-azurin fusion protein on breast cancer cells. Mole Biol Rep 46: 3129-3140. [Crossref]
- Wang T, Zhao J, Ren JL, Zhang L, Wen WH et al. (2007) Recombinant immunoproapoptotic proteins with furin site can translocate and kill HER2-positive cancer cells. Cancer Res 67: 11830-11839. [Crossref]
- Nikkhoi SK, Rahbarizadeh F, Ranjbar S, Khaleghi S, Farasat A (2018) Liposomal nanoparticle armed with bivalent bispecific single-domain antibodies, novel weapon in HER2 positive cancerous cell lines targeting. Mole Immunol 96: 98-109. [Crossref]
- Medema JP, de Jong J, Peltenburg L, Verdegaal E, Gorter A et al. (2001) Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc Nat Acad Sci 98: 11515-11520. [Crossref]
- Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69: 7-34. [Crossref]
- Lezhnin YN, Kravchenko YE, Frolova EI, Chumakov PM, Chumakov SP (2015) Oncotoxic proteins in cancer therapy: Mechanisms of action. Mole Biol 49: 231-243. [Crossref]
- de Goeij BE, Lambert JM (2016) New developments for antibody-drug conjugate-based therapeutic approaches. Curr Opin Immunol 40: 14-23. [Crossref]
- Bar-Zeev M, Livney YD, Assaraf YG (2017) Targeted nanomedicine for cancer therapeutics: Towards precision medicine overcoming drug resistance. Drug Resist Updates 31: 15-30. [Crossref]
- Mehta RR, Hawthorne M, Peng X, Shilkaitis A, Mehta RG et al. (2010) A 28-amino-acid peptide fragment of the cupredoxin azurin prevents carcinogen-induced mouse mammary lesions. Cancer Prev Res 3: 1351-1360. [Crossref]
- Schiffer S, Hansen H, Hehmann-Titt G, Huhn M, Fischer R et al. (2013) Efficacy of an adapted granzyme B-based anti-CD30 cytolytic fusion protein against PI-9-positive classical Hodgkin lymphoma cells in a murine model. Blood Cancer J 3: e106. [Crossref]
- Losasso V, Schiffer S, Barth S, Carloni P (2012) Design of human granzyme B variants resistant to serpin B9. Proteins 80: 2514-2522. [Crossref]
- Jabulowsky RA, Oberoi P, Bähr-Mahmud H, Dälken B, Wels WS (2012) Surface Charge-Modification Prevents Sequestration and Enhances Tumor-Cell Specificity of a Recombinant Granzyme B–TGFα Fusion Protein. Bioconjug Chem 23: 1567-1576. [Crossref]
- Wang F, Ren J, Qiu XC, Wang LF, Zhu Q et al. (2010) Selective cytotoxicity to HER2-positive tumor cells by a recombinant e23sFv-TD-tBID protein containing a furin cleavage sequence. Clinical Cancer Res 16: 2284-2294. [Crossref]
- Goyal A, Batra JK (2000) Inclusion of a furin-sensitive spacer enhances the cytotoxicity of ribotoxin restrictocin containing recombinant single-chain immunotoxins. Biochem J 345: 247-254. [Crossref]
- Chen X, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65: 1357-1369. [Crossref]
- Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A et al. (1998) Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity 8: 451-460. [Crossref]
- Rousalova I, Krepela E (2010) Granzyme B-induced apoptosis in cancer cells and its regulation. Int J Oncol 37: 1361-1378. [Crossref]
- Kurschus FC, Kleinschmidt M, Fellows E, Dornmair K, Rudolph R et al. (2004) Killing of target cells by redirected granzyme B in the absence of perforin. FEBS Lett 562: 87-92. [Crossref]
