In silico Analysis of Alkaloids for Therapeutic Use in Treatment of Alzheimer's Disease by Targeting Acetylcholinesterase Enzyme
A B S T R A C T
Alzheimer’s is a progressive mental deterioration associated with the degeneration of the cognition activities and memory loss. It is considered to be a multifactorial disease. One of the causes of the Alzheimer’s disease is the low concentration of the neurotransmitter named acetylcholine (ACh) at the synaptic cleft. Thus, inhibitor of Acetylcholinesterase (AChE), an enzyme whose function is to degrade the acetylcholine, is proved to be a promising candidate to treat this disease. Among the inhibitors are the natural alkaloids that also have an inhibitory effect on the AChE. In this study we have focused on the simple derivates of beta carbolime (a group of alkaloids) and studied their interaction with AChE via rigid protein-ligand docking approach.
Keywords
Beta carbolines, alkaloids, docking, acetylcholinesterase, inhibitor
Introduction
Alzheimer’s disease is a progressive, age related neurodegenerative disorder that affects nearly 2% of the population of industrialized countries [1]. It is generally diagnosed in individuals above 65 years of age and is the primary cause of the memory loss (dementia) in elderly individuals. Each year 4.6 million new cases of Alzheimer’s disease are reported and according to a report generated by World Health Organization (WHO), by 2040, 71% of the 81.1 million dementia cases will occur in developing countries [2, 3]. Alzheimer’s disease is considered to be a multifactorial disease i.e., having more than one factors contributing to the pathogenesis of the disease. It is caused by the accumulation of amyloid beta. Amyloid beta is a peptide of 36 to 43 amino acids which is formed as a result of the cleavage of amyloid beta precursor protein by beta secretase and gama secretase enzyme [4]. Amyloid beta starts up as a single molecule which initially clusters up to form the plagues. As a result, the connection between the synapses is disrupted which causes the loss of memory and other cognitive abilities [5].
Another factor contributing to Alzheimer’s disease is the low concentration of ACh at the synaptic cleft resulting in the death of the cholinergic neurons which causes the loss of the memory [6]. Acetylcholinesterase (AChE) is an enzyme present in the nervous tissue, muscles and red blood cells that hydrolyze acetylcholine into choline and acetic acid. Thus the inhibition of the acetylcholinesterase enzyme will prevent the breakdown of the acetylcholine present in the nerve cells and will result in the increase of the Ach concentrations thus preventing the Alzheimer’s disease (and memory loss) [7]. The physiological importance of the ACh enhanced the focus on the inhibitors of acetylcholinesterase. Tetrohydroaminoacridine (THA) named as Tacrine or Cognex was the first drug approved by US FDA (Food and Drug administration) in 1993 for the treatment of Alzheimer’s disease. Its use improved the memory and cognitive ability of Alzheimer’s patients but had a lot of side effects including nausea, liver dysfunction, vomiting, diarrhea and dizziness. Another drug Donepezil was approved by FDA in 1996 having more substrate specifity thus minimizing the side effects. Rivastigmine, galantamine, memantine were approved by FDA in 2000, 2001 and 2003 respectively. All of these drugs work by inhibiting acetylcholinesterase thus improving ACh concentration in synaptic cleft and as a result memory and other cognitive abilities are improved [8, 9].
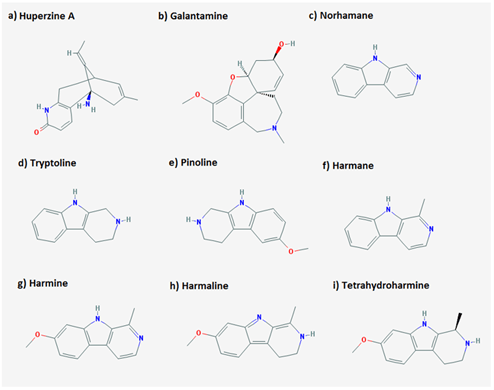
Among the natural products, alkaloids are considered to be the most promising candidates for the inhibition of acetylcholinesterase, consequently it can be used in the treatment of the Alzheimer’s disease [10]. We focused on the simple derivatives of beta carboline, a subcategory of alkaloids. Beta carboline belongs to a group of indole alkaloids and consist of pyridine ring that is fused to an indole skeleton. In this study, we listed down the simple derivatives of beta carboline (Huperzine A, Galantamine, norharmane, Tryptoline, Pinoline, Harmane, Harmine, Harmaline) and performed in silico analysis to identify their interactions with acetylcholinesterase. The crystal structure of acetylcholinesterase complexed with Huperzine A is available on Protein databank (PDB) [11]. Huperzine A is an alkaloid possessing acetylcholine inhibition properties. Thus, this complex was taken as a reference and the interacting residues were correlated with our results.
Materials and Methods
I Receptor and Ligand Dataset
The crystal structure of acetylcholinesterase complexed with Huperzine A (PDB ID: 4EY5) was taken as a reference and was retrieved from Protein Data Bank (PDB) [11]. The binding site residues of this complex were extracted via Ligplot [12]. Acetylcholinesterase was detached from Huperzine A and again re docked to verify the interacting residues. The residues involved in the interaction of crystallized structure of AChE with Huperzine A was same as that of the residues obtained after docking. Table 1 list down the binding residues of acetylcholinesterase with Huperzine A. 3D Structures of ligands were extracted from PubChem database [13]. Figure 1 shows the simple derivatives of beta carboline alkaloids used in this study as ligands.
Table 1: Interacting residues of acetylcholinesterase.
|
Binding residues of Acetylcholinesterase |
|
Tyr133, Tyr119, Gly120, Gly126, Gly122, Ser125, Tyr337, Gly121, Tyr124,
His447, Trp86, Ser203, Glu202 |
Figure 1: 2D structure of ligands used in the study.
II Docking Studies
Molecular dockings are performed to predict the binding orientation between a receptor and its ligand to form a stable molecular complex [14]. Docking allows having a detailed insight into the interacting residues in the form of 3D structure. Docking studies also allows for the identification of the drug candidates and for designing novel or new effective drugs [15]. In this study rigid docking was performed i.e., the receptor undertaken in this study was studied in its static state rather than the dynamic state. Acetylcholinesterase was docked against all the ligands using Autodock 4.2. Autodock is a suite of automated docking programmes and it is designed to predict how substrates or drugs bind with the receptor in the best possible conformation having lowest energy thus making the structure stable [16]. A total run of acetylcholinesterase with each respective ligand was set to 50 i.e., the ligand binds to its receptor in 50 different conformations and orientations. The grid size was set to cover complete receptor thus allowing more space for the ligand to bind i.e., the ligand can bind anywhere on the receptor (blind docking). Ideally, the ligands were supposed to occupy the same bind site as that of the Huperzine A with AChE. Table 2 shows the detailed docking parameters used. The docking results were analysed using Autodock tools 4.2 [17]. The energy values as well as interacting residues were noted down. The 2D and 3D interactions of the complexes were visualized in Discovery Studio 4.2 [18]. The 2D interactions provide insight into the hydrogen bonds as well as hydrophobic interactions. These interactions were highlighted in 3D structure to gain a deeper insight of the atoms of specific residues involved in interactions and contributing towards the stability of the complex.
Table 2: Autodock 4.2 parameters used for docking.
|
Grid Parameters |
Docking Parameters |
||
|
Spacing |
0.375Å |
Energy evaluations |
2.5 x 106 |
|
Grid Center |
80X Å 80Y Å 80Z Å |
Iterations |
27000 |
|
Mutation rate |
0.02 |
||
|
Crossover rate |
0.80 |
||
|
|
|
Elitism value |
1 |
|
|
|
RMS Tolerance |
1.0 Å |
Results and Discussions
Molecular dockings were performed on acetylcholinesterase receptor with the ligand dataset using Autodock 4.2, to identify the binding residues involved for each ligand in interaction with the receptor. The docking conformations were run based on the energy values; thus, the first structure corresponds to the lowest energy conformation. The overall stability of a complex is directly associated to the free energy of that complex. Lower energy values suggest the presence of a high binding affinity between the ligand and receptor. It is of substantial importance to highlight each complex’s free energy in order to evaluate the successful complex formation. Free energy or unbound energy should be zero thus representing that all atoms present within the residues are bonded correctly with respect to their valancies. Respective energy values of all the ligands are shown in (Table 3). The binding energy values and the ligand efficiency for all the complexes fall below the value of 0. This suggests the formation of relatively stable complexes between acetylcholinesterase receptor with all the ligands. Each complex was then analysed on Ligplot to identify the binding residues of the receptor. Detailed analysis of the binding site conformations of each ligand showed that all the ligands bind with the conserved binding site of acetylcholinesterase receptor. The residues were same as those of acetylcholinesterase with Huperzine A thus validating our docking studies.
Table 3: Energy values of the ligands.
|
Ligands |
Binding Energy |
Ligand Efficiency |
Inhib Constant (µM) |
Intermol Energy |
Vdw HB desolv Energy |
Electrostatic Energy |
Torsional Energy |
Unbound Energy |
|
Huperzine
A |
-8.69 |
-0.48 |
427.57 |
-8.99 |
-7.59 |
-1.4 |
0.3 |
0.0 |
|
Galantamine |
-8.42 |
-0.4 |
678.11 |
-9.01 |
-8.15 |
-0.87 |
0.6 |
0.0 |
|
norharmane |
-6.62 |
-0.51 |
14.03 |
-6.62 |
-6.59 |
-0.03 |
0.0 |
0.0 |
|
Tryptoline |
-6.45 |
-0.5 |
18.83 |
-6.45 |
-6.37 |
-0.07 |
0.0 |
0.0 |
|
Pinoline |
-6.61 |
-0.44 |
14.33 |
-6.91 |
-6.74 |
-0.16 |
0.3 |
0.0 |
|
Harmane |
-6.99 |
-0.5 |
7.56 |
-6.99 |
-6.95 |
-0.04 |
0.0 |
0.0 |
|
Harmine |
-7.17 |
-0.45 |
5.59 |
-7.46 |
-7.52 |
0.05 |
0.3 |
0.0 |
|
Harmaline |
-6.79 |
-0.42 |
10.6 |
-7.09 |
-7.05 |
-0.04 |
0.3 |
0.0 |
|
Tetrahydroharmine |
-6.91 |
-0.43 |
8.6 |
-7.21 |
-7.15 |
-0.05 |
0.3 |
0.0 |
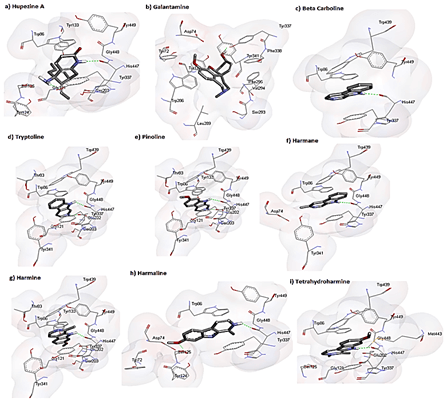
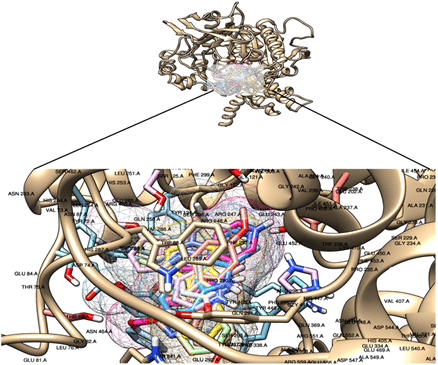
Table 4 shows the detailed 2D analysis retrieved from plotting the binding interaction residues on Discovery studio and Ligplot. Residue information for each complex including H-Bond distances, atoms involved in the formation of H-bonds and residues taking part in the hydrophobic interactions are shown in (Table 4). The 3D pictorial representation of the interactions among acetylcholinesterase with its ligands is shown in (Figure 2). Hydrogen bonds are represented via green dotted lines. Each ligand occupies the same binding site on Acetylcholinesterase in its own respective orientation to form the most stable complex. However, each ligand is shown to bind to the same binding residues as reported in literature. After analysing each complex individually all the complexes were superimposed using chimera in order to determine whether they occupy the same position or not. The binding pocket of Acetylcholinesterase with ligands is shown in (Figure 3). It is shown clearly that all the ligands occupied the same binding pocket in structure of Acetylcholinesterase. The binding residues were the same residues that have been reported previously with Huperzine A.
Table 4: Docking results of acetylcholinesterase with respective ligands.
|
Ligand |
Hydrogen Bond |
Atoms Involved |
Distance (Å) |
Hydrophobic Interacting
Residues |
|
Huperzine A |
Tyr337 His447 |
O-NH O-NH |
1.8 1.8 |
Tyr124, Trp86, Tyr133 |
|
Galantamine |
Tyr337 |
N-OH |
2.2 |
Asp74, Phe295, Tyr124, Trp286, Phe338, Val294, Tyr341, Ser293, Arg296,
Tyr72 |
|
β-carboline |
His447 |
O-N1 |
2.77 |
Trp86, Trp439, Tyr337, Tyr449 |
|
Tryptoline |
His447 Glu202 |
O-N1 OE1-N2 |
2.55 3.17 |
Gly121, Tyr337, Trp86, Trp439, Gly448 |
|
Pinoline |
His447 Glu202 |
O-N1 OE1-N2 |
2.76 2.71 |
Ser203, Gly121, Trp86, Gly448, Tyr337, Trp439 |
|
Harmane |
His447 |
O-N1 |
2.77 |
Trp86, Tyr449, Tyr337, Trp439, Pro446 |
|
Harmine |
His447 |
O-N1 |
2.67 |
Trp86, Tyr337, Gly121, Glu202, Trp439, Tyr449, Gly448, Pro446 |
|
Harmaline |
His447 Ser125 |
O-N1 O16-OG |
2.83 2.78 |
Trp86, Asp74, Tyr124, Try337, Gly448, Tyr72 |
|
Tetrahydroharmine |
His447 |
O-N2 |
2.76 |
Trp86, Tyr337, Tyr449, Met443, Trp439, Pro446 |
Figure 2: 3D representation of interactions among acetylcholinesterase and respective ligands. receptor is shown in surface while ligands are shown in sticks. Hydrogen bonds are represented in green dotted lines.
Figure 3: Overall binding pattern of all ligands in acetylcholinesterase pocket.
Conservation of Acetylcholinesterase
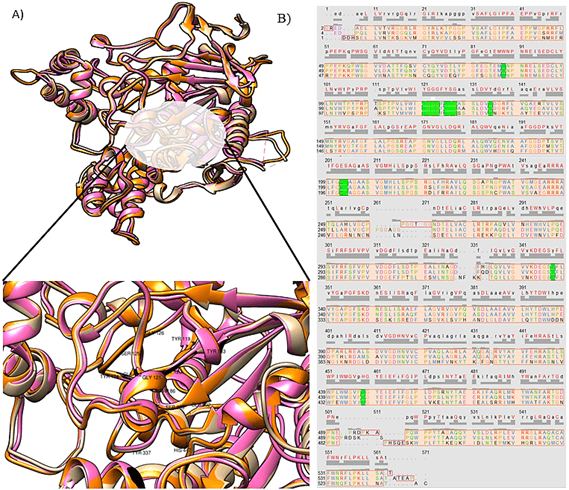
In order to check the conservation of the acetylcholinesterase binding site across the species with alkaloids, we retrieved the pdbs for human AChE bound with Huperzine A, AChE of pacific electric ray Torpedo californica bound with Huperzine A and mouse AChE bound with huprine [19-21]. The three structures were superimposed upon each other using chimera and were aligned structurally and sequentially. The binding pocket residues were checked for the conservation across three species. The conservation of the binding pocket highlights the binding interaction between the receptor and ligand thus making it an important target for drug designing across the species. The structure and sequence of the binding pocket of human and mouse AChE with Huperzine A is more conserved as compared to AChE of Torpedo californica bound to Huperzine A. Figure 4A shows the superimposition of the AChE of three species. The conserved residues of the binding site are labeled. Figure 4B shows the cross-species sequence alignment of AChE. The conserved residues are highlighted with green colour.
Figure 4: A) Superimposition of AChE of human, mouse and electric ray bound with Huperzine A. Beige colour represents AChE for human, pink represents mouse AChE, orange represents fish AChE. Conserved binding residues are labeled. B) Cross species sequence alignment of AChE. Conserved binding site residues are highlighted in green colour.
Conclusion
AChE, an enzyme involved in the breakdown of the neurotransmitter ACh, has been in focus for the treatment of the cognitive impairment associated with Alzheimer’s by using inhibitors of this enzyme. Beta carboline alkaloids have been reported to act as an inhibitor of AChE. In this study we have performed an in silico analysis of binding interactions of beta carboline alkaloids with AChE. Docking studies were performed to study the interaction and to identify the residues in the binding pocket occupied by each of the beta carboline alkaloids. The binding mode of each interaction was similar to that of the reported for Huperzine A. Moreover, the conservation of the binding pocket among mouse, human and electric ray fish was also checked.
Article Info
Article Type
Research ArticlePublication history
Received: Tue 04, May 2021Accepted: Fri 14, May 2021
Published: Fri 28, May 2021
Copyright
© 2023 Sidra Batool. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.NNB.2021.02.02
Author Info
Muhammad Sibte Hasan Mahmood Tiyyaba Furqan Muaaz Karim Sidra Batool
Corresponding Author
Sidra BatoolDepartment of Biosciences, COMSATS University, Chak Shahzad, Islamabad, Pakistan
Figures & Tables
Table 1: Interacting residues of acetylcholinesterase.
|
Binding residues of Acetylcholinesterase |
|
Tyr133, Tyr119, Gly120, Gly126, Gly122, Ser125, Tyr337, Gly121, Tyr124,
His447, Trp86, Ser203, Glu202 |
Table 2: Autodock 4.2 parameters used for docking.
|
Grid Parameters |
Docking Parameters |
||
|
Spacing |
0.375Å |
Energy evaluations |
2.5 x 106 |
|
Grid Center |
80X Å 80Y Å 80Z Å |
Iterations |
27000 |
|
Mutation rate |
0.02 |
||
|
Crossover rate |
0.80 |
||
|
|
|
Elitism value |
1 |
|
|
|
RMS Tolerance |
1.0 Å |
Table 3: Energy values of the ligands.
|
Ligands |
Binding Energy |
Ligand Efficiency |
Inhib Constant (µM) |
Intermol Energy |
Vdw HB desolv Energy |
Electrostatic Energy |
Torsional Energy |
Unbound Energy |
|
Huperzine
A |
-8.69 |
-0.48 |
427.57 |
-8.99 |
-7.59 |
-1.4 |
0.3 |
0.0 |
|
Galantamine |
-8.42 |
-0.4 |
678.11 |
-9.01 |
-8.15 |
-0.87 |
0.6 |
0.0 |
|
norharmane |
-6.62 |
-0.51 |
14.03 |
-6.62 |
-6.59 |
-0.03 |
0.0 |
0.0 |
|
Tryptoline |
-6.45 |
-0.5 |
18.83 |
-6.45 |
-6.37 |
-0.07 |
0.0 |
0.0 |
|
Pinoline |
-6.61 |
-0.44 |
14.33 |
-6.91 |
-6.74 |
-0.16 |
0.3 |
0.0 |
|
Harmane |
-6.99 |
-0.5 |
7.56 |
-6.99 |
-6.95 |
-0.04 |
0.0 |
0.0 |
|
Harmine |
-7.17 |
-0.45 |
5.59 |
-7.46 |
-7.52 |
0.05 |
0.3 |
0.0 |
|
Harmaline |
-6.79 |
-0.42 |
10.6 |
-7.09 |
-7.05 |
-0.04 |
0.3 |
0.0 |
|
Tetrahydroharmine |
-6.91 |
-0.43 |
8.6 |
-7.21 |
-7.15 |
-0.05 |
0.3 |
0.0 |
Table 4: Docking results of acetylcholinesterase with respective ligands.
|
Ligand |
Hydrogen Bond |
Atoms Involved |
Distance (Å) |
Hydrophobic Interacting
Residues |
|
Huperzine A |
Tyr337 His447 |
O-NH O-NH |
1.8 1.8 |
Tyr124, Trp86, Tyr133 |
|
Galantamine |
Tyr337 |
N-OH |
2.2 |
Asp74, Phe295, Tyr124, Trp286, Phe338, Val294, Tyr341, Ser293, Arg296,
Tyr72 |
|
β-carboline |
His447 |
O-N1 |
2.77 |
Trp86, Trp439, Tyr337, Tyr449 |
|
Tryptoline |
His447 Glu202 |
O-N1 OE1-N2 |
2.55 3.17 |
Gly121, Tyr337, Trp86, Trp439, Gly448 |
|
Pinoline |
His447 Glu202 |
O-N1 OE1-N2 |
2.76 2.71 |
Ser203, Gly121, Trp86, Gly448, Tyr337, Trp439 |
|
Harmane |
His447 |
O-N1 |
2.77 |
Trp86, Tyr449, Tyr337, Trp439, Pro446 |
|
Harmine |
His447 |
O-N1 |
2.67 |
Trp86, Tyr337, Gly121, Glu202, Trp439, Tyr449, Gly448, Pro446 |
|
Harmaline |
His447 Ser125 |
O-N1 O16-OG |
2.83 2.78 |
Trp86, Asp74, Tyr124, Try337, Gly448, Tyr72 |
|
Tetrahydroharmine |
His447 |
O-N2 |
2.76 |
Trp86, Tyr337, Tyr449, Met443, Trp439, Pro446 |
References
1.
Mattson MP (2004) Pathways towards and away from Alzheimer’s disease.
Nature 430: 631-639. [Crossref]
2.
Ferri CP, Prince M, Brayne C,
Brodaty H, Fratiglioni L et al. (2005) Global prevalence of dementia: a Delphi
consensus study. Lancet 366: 2112-2117. [Crossref]
3.
Kalaria RN, Maestre GE, Arizaga R, Friedland RP, Galasko D et al. (2008)
Alzheimer’s disease and vascular dementia in developing countries: prevalence,
management, and risk factors. Lancet Neurol 7: 812-826. [Crossref]
4.
Murphy MP, LeVine H (2010) Alzheimer’s Disease and the β-Amyloid Peptide.
J Alzheimers Dis 19: 311. [Crossref]
5.
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease:
progress and problems on the road to therapeutics. Science 297: 353-356.
[Crossref]
6.
Perry E (1988) Acetylcholine and Alzheimer’s Disease. Br J Psychiatry
152: 737-740. [Crossref]
7.
Soreq H, Seidman S (2001) Acetylcholinesterase--new roles for an old
actor. Nat Rev Neurosci 2: 294-302. [Crossref]
8.
Hansen RA, Gartlehner G, Webb AP, Morgan LC, Moore CG et al. (2008)
Efficacy and safety of donepezil, galantamine, and rivastigmine for the
treatment of Alzheimer’s disease: A systematic review and meta-analysis. Clin
Interv Aging 3: 211-225. [Crossref]
9.
Scarpini E, Schelterns P, Feldman H (2003) Treatment of Alzheimer’s
disease; current status and new perspectives. Lancet Neurol 2: 539-547.
[Crossref]
10.
Pereira DM, Ferreres F,
Oliveira JMA, Gaspar L, Faria J et al. (2010) Pharmacological effects of Catharanthus
roseus root alkaloids in acetylcholinesterase inhibition and cholinergic
neurotransmission. Phytomedicine 17: 646-652. [Crossref]
11.
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD et al. (1978)
The Protein Data Bank: a computer-based archival file for macromolecular
structures. Arch Biochem Biophys 185: 584-591. [Crossref]
12.
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to
generate schematic diagrams of protein-ligand interactions. Protein Eng
8: 127-134. [Crossref]
13.
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J et al. (2009) PubChem: a public
information system for analyzing bioactivities of small molecules. Nucleic
Acids Res 37: 623-633. [Crossref]
14.
Schulz GE (1992) Binding of nucleotides by proteins: Current opinion in
structural biology 1992, 2: 61…-67. Curr Opin Struc Bio 2: 61-67.
15.
Mondal C, Halder AK, Adhikari N, Saha A, Das Saha K et al. (2015)
Comparative validated molecular modeling of p53-HDM2 inhibitors as
antiproliferative agents. Eur J Med Chem 90: 860-875. [Crossref]
16.
Morris GM, Huey R, Olson AJ (2008) Using AutoDock for ligand-receptor
docking. Curr Protoc Bioinformatics 8: 14. [Crossref]
17.
Morris G, Huey R, Lindstrom W, Sanner M, Belew R et al. (2009) AutoDock4
and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J
Comp Chem 30: 2785-2791.
18.
Visualizer DS (2005) Accelrys Software Inc. Disc Stud Visua.
19.
Cheung J, Rudolph MJ, Burshteyn F, Cassidy MS, Gary EN et al. (2012)
Structures of human acetylcholinesterase in complex with pharmacologically
important ligands. J Med Chem 55: 10282-10286. [Crossref]
20.
Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP et al. (1997)
Structure of acetylcholinesterase complexed with the nootropic alkaloid,
(–)-huperzine A. Nat Struct Biol 4: 57-63. [Crossref]
21. Ronco
C, Carletti E, Colletier JP, Weik M, Nachon F et al. (2012)
Huprine derivatives as sub-nanomolar human acetylcholinesterase inhibitors:
from rational design to validation by X-ray crystallography. ChemMedChem
7: 400-405. [Crossref]
