Journals
Gene expression profiling in hepatoblastoma cases of the Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) trial
A B S T R A C T
Background:Over the past two decades, significant improvements in the outcomes of children diagnosed with hepatoblastoma (HBL) have resulted from developments in diagnostic methods and treatments. However, the outcomes of some cases remain unfavorable, and others suffer from late complications caused by treatment. Procedure:We elucidated the genetic profile of HBLs to identify biological markers for diagnosis and to determine the grade of malignancy. RNA samples extracted from 53 specimens of fresh-frozen HBL and corresponding noncancerous liver tissues were used for gene expression profiling and pathway analysis. Results:In the comparison between HBL and noncancerous liver tissues, genes involved in several transcription pathways including glypican 3 and Wnt signaling pathway members were upregulated, whereas cytochrome p450 family genes were downregulated. The analyses of high-risk (metastatic) HBL and HBL progression or recurrence cases revealed upregulated expression of histone cluster genes and upregulation of RXR activators molecular signaling. Clustering analysis of the tumor samples revealed three distinct groups among the HBL cases. Conclusion:Aberrant expression of genes involved in tissue differentiation pathways may be related to the development of HBL, and such genes, including AFP, PGC, SPINK1, and NQO1, may be putative therapeutic targets for HBL progression.
Keywords
Hepatoblastoma,Microarray,gene expression,clustering,malignancy grade
Introduction
Hepatoblastoma (HBL) is the most common primary liver tumor in children and is usually diagnosed during the first 3 years of life. Before 1980, the only curative treatment for children with malignant hepatic tumors was complete surgical resection of the tumor. The introduction of effective chemotherapeutic regimens in the 1980s resulted in an increased number of patients ultimately eligible for tumor resection and a reduced postoperative recurrence rate [1-3]. However, poor outcomes are still seen in several HBL cases, especially those with distant metastasis. The identification and development of new prognostic stratifications have led to novel treatments for high-risk patients and reduced treatment for low-risk patients to avoid the delayed effects and unnecessary toxicities associated with treatment [4]. Until now, few biological markers to stratify high-risk tumors have been reported, and no biological markers have been applied in the clinic [5, 6]. To identify new molecular biomarkers for risk stratification and malignant grade, we performed microarray analysis of gene expression in samples from HBL patients prior to treatment.
Materials and Methods
I Patients
Among approximately 400 patients enrolled in Japanese Study Group for Pediatric Liver Tumors -2 (JPLT-2) trial, approximately 360 hepatoblastoma (HBL) patients underwent this protocol between December 1999 and November 2012 at the institutions of the JPLT. Tumor and noncancerous liver tissue (NCL) samples from more than 100 of these HB patients were obtained at diagnosis before chemotherapy and stored at -80?C. The JPLT-2 consisted of two different treatment protocols: cisplatin and pirarubicin as first-line treatment and ifosfamide, pirarubicin, etoposide, and carboplatin as second-line treatment [7]. The human ethics review committee of our university approved the study protocol, and signed informed consent was obtained from the parent of each patient (Ethics Committee Approval No. 20). The clinical disease stage was determined at the time of initial diagnosis according to the classification of the pre-treatment extent of disease (PRETEXT) staging system, which is based on the number of liver segments involved, extent of local invasion, extent of regional lymph node involvement, and presence of distant metastasis [8]. The Childhood Hepatic Tumors International Collaboration (CHIC) is a new international hepatoblastoma database created by four major study groups: the International Childhood Liver Tumors Strategy Group (SIOPEL), Children's Oncology Group, German Association of Pediatric Hematology and Oncology, and JPLT [9, 10]. Risk stratification was also performed according to the CHIC classification criteria based on clinical backbones. Briefly, standard-risk disease is defined as PRETEXT I/II tumors or PRETEXT III tumors with negative PRETEXT annotation factors, intermediate-risk disease as PRETEXT IV tumors or PRETEXT I -III tumors with positive PRETEXT annotation factors, and high-risk disease as metastatic disease. The pathological classification of HBLs by Haas et al. and the Japanese Society of Pathology grouped HBLs into two major subtypes: well-differentiated (fetal) and poorly differentiated (embryonal) types [8, 11]. In this study, we enrolled 53 patients whose tumor RNA quality was sufficient for microarray analysis. The clinical features of the patients are summarized in (Table 1).
II Tissue samples
Tumor tissue were obtained during surgery or biopsy from more than 100 patients prior to any chemotherapy and corresponding noncancerous liver (NCL) tissue specimens were also obtained to the extent possible. Among the 53 cases whose tumor RNA was high quality, 14 NCL samples were available. These samples were immediately stored at ?80 ºC until use. The tissue adjacent to each frozen tissue specimen was examined by pathological review for diagnosis and confirmation.
III DNA and RNA extraction
Tissue DNA samples were extracted and purified using standard methods. Total cellular RNA was extracted from tumor tissues by the acid guanidinium thiocyanate -phenol -chloroform method [12]. The quantity and quality of the extracted RNA were assessed using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA, USA). We selected 53 RNA samples whose RNA integrity number (RIN) values were greater than 7.4.
IV Detection of CTNNB1 (Catenin Beta 1) gene mutations and deletions
To detect mutations and deletions in CTNNB1, genomic DNA from each tumor specimen and corresponding noncancerous liver tissue was amplified by PCR using primers targeting exon 3 of CTNNB1, as described previously [13]. To detect amplicons harboring a large deletion event involving exon 3, the sizes of the PCR products were analyzed by 2% agarose gel electrophoresis. To detect point mutations in exon 3, the PCR products were reamplified using the following internal primers: 5'-AAAATCCAGCGTGGACAATGG-3 and 5'-TGTGGCAAGTTCTGCATCATC-3'. The resulting PCR products were sequenced using the ABI 3100 DNA sequences. Mutations were confirmed by at least two amplification reactions from the original DNA.
V Microarray analysis
Microarray experiments were performed using the Affymetrix Gene Chip (Affymetrix, Inc., Santa Clara, CA, USA), according to the standard protocols, and the Ambion WT Expression kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). First-strand cDNA was generated from 250 ng total RNA obtained from HB tumor and normal liver tissues using reverse transcriptase and the T7 primer. Second-strand cDNA was generated using a DNA polymerase mix containing RNase H. Antisense complementary RNA (cRNA) was generated via in vitro transcription using the T7 RNA polymerase and was purified using nucleic-acid-binding beads. Second-cycle cDNA was generated from the purified antisense cRNA; 5.5 µg of the resulting cDNA were then subjected to fragmentation and terminal labeling and hybridized to the Human Gene 1.0 ST array (Affimetrix, Thermo Fisher Scientific, Santa Clara, CA), which contains 36,079 probes including 32,020 annotated RefSeq transcripts. After hybridization, the arrays were rinsed and labeled with streptavidin-Cy5, scanned using a scanner (GeneChip™ Scanner 3000 7G, Affimetrix, Thermo Fisher Scientific), and then analyzed using Gene Spring Software (Gene Spring GX ver. 14.9, Agilnet, Santa Clara, CA). The microarray data were registered in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov.geo/).
VI Microarray data analysis
For comparisons of gene expression fold changes determined from the microarray data, we considered the RefSeq probes on the array to be nonredundant and representative of the whole transcriptome. First, we compared gene expression changes between HBL and NCL tissues. In the 14 cases with available tumor and NCL tissues, gene expression levels between paired tumor and NCL tissues were performed. Otherwise, gene expression levels were also compared between whole expression data of 53 tumor samples and that of 14 noncancerous liver tissue. Second, among the HBL samples, we compared gene expression levels among the standard-, intermediate-, and high-risk HBL cases. Gene expression between the event-free surviving and recurrence or progression cases was also compared. The resulting gene lists were filtered and uploaded to the Ingenuity Pathway Analysis (TOMY Digital Biology, Inc., Tokyo, Japan) and Key Molnet ver. 2 (KM Data Co., Tokyo, Japan) software programs.
Table 1. Clinicopathological features of the hepatoblastoma samples used for microarray analysis.
|
Age (months) |
|
|
Median |
22 |
|
Range |
0-109 |
|
Sex |
|
|
Female |
25 (47.2%) |
|
Male |
28 (52.8%) |
|
Histologic type |
|
|
Well differentiated |
30 (56.6%) |
|
Poorly differentiated |
21 (39.6%) |
|
Other |
2 (3.8%) |
|
PRETEXT classificationa |
|
|
I |
9 (18.0%) |
|
II |
15 (28.3%) |
|
III |
18 (34.0%) |
|
IV |
11 (20.7%) |
|
Distant metastasis |
14 (26.4%) |
|
CHIC risk stratificationb |
|
|
Standard |
31 (58.5%) |
|
Intermediate |
8 (15.1%) |
|
High |
14 (26.4%) |
|
CTNNBI gene alteration |
|
|
Deletion |
23 (43.4%) |
|
Mutation (exon 3) |
16 (18.9%) |
|
Event |
|
|
Free |
32 (60.4%) |
|
+ |
21 (39.6%) |
|
Clinical course |
|
|
Alive |
38 (71.7%) |
|
Dead |
15 (28.3%) |
aPRETEXT: PRETreatment of EXTent of disease 2,7, bCHIC: Children Hepatic tumor International Collaboration 9,10
VII Statistical analysis
The Mann-Whitney U test and Chi-square test were used to examine the relationships between clinical factors and gene expression profiles. All tests were two tailed, and differences with a P-value < 0.05 were regarded as statistically significant.
Results
I Gene expression profiles of HBL tissue compared with noncancerous liver tissue
The gene expression profiles of noncancerous liver(NCL)and HBL tissues were obviously different. Therefore, the differences in expression of specific genes between the tumor and NCL tissue samples were analyzed. The lists of the upregulated and downregulated genes are shown in Table 2. The upregulated genes were those with a greater than 10-fold increase in expression in more than half of HBL samples compared with the paired NCL tissues, and the downregulated genes were those with a less than 15-fold decrease in expression in more than half of HBL samples. Of these genes, REG3A (Regenerating Family Member 3 Alpha), glypican 3 (GPC3), and Wnt signaling-related genes were upregulated in HBL. The network analysis of differentially expressed genes encoding transcription factors, including HNF1A (Hepatocyte Nuclear Factor 1 Alpha), HNF4A (Hepatocyte Nuclear Factor 4 Alpha), C/EBP, CTNNB1, and FOXA3, indicated several pathways including FXR (farnesoid X receptor) /RXR (retinoid X receptor activation), LXR (liver X receptor) /RXR and PXR (pregnane X receptor)/RXR pathways potentially related to tumor development [14]. The downregulated genes (Table 2) included cytochrome P450 (CYP) family genes, solute carrier family genes, and genes encoding enzymes, such as tyrosine aminotransferase and tryptophan 2,3-dioxygenase. Of the CYP family genes, CYP2B6, CYP2C8, CYP2B7P1, and CYP2B7P1 were markedly downregulated. The pathway analysis indicated upstream genes including PXR and CAR are suspected as regulatory factors in these genes. The network analysis of differentially expressed genes also indicated genes associated with complement activation and blood coagulation.
Table 2: Up- and downregulated genes in hepatoblastoma tissues compared with noncancerous tissues
|
Gene symbol |
Gene |
Location |
|
Upregulated genes |
||
|
REG3A |
regenerating islet-derived 3 alpha |
chr2 |
|
DPEP1 |
dipeptidase 1 (renal) |
chr16 |
|
SNORD114-26 |
small nucleolar RNA, C/D box 114-26 |
chr14 |
|
LGR5 |
leucine-rich repeat-containing G protein-coupled receptor 5 |
chr12 |
|
DKK1 |
dickkopf homolog 1 (Xenopus laevis) |
chr10 |
|
EPCAM |
epithelial cell adhesion molecule |
chr4 |
|
TSPAN5 |
tetraspanin 5 |
chr4 |
|
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
chr18 |
|
SNORD113-4 |
small nucleolar RNA, C/D box 113-4 |
chr14 |
|
NOTUM |
notum pectinacetylesterase homolog (Drosophila) |
chr17 |
|
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
chr12 |
|
NKD1 |
naked cuticle homolog 1 (Drosophila) |
chr16 |
|
DKK4 |
dickkopf homolog 4 (Xenopus laevis) |
chr8 |
|
MEP1A |
meprin A, alpha (PABA peptid e hydrolase) |
chr6 |
|
SLC7A11 |
solute carrier family 7, member 11 |
chr4 |
|
PEG10 |
paternally expressed 10 |
chr7 |
|
DLK1 |
delta-like 1 homolog (Drosophila) |
chr14 |
|
C9orf4 |
chromosome 9 open reading frame 4 |
chr9 |
|
GPC3 |
glypican 3 |
chrX |
|
Downregulated genes |
||
|
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
chr19 |
|
CYP2C8|CYP2C19 |
cytochrome P450, family 2, subfamily C, polypeptide 8 | 19 |
chr10 |
|
TAT |
tyrosine aminotransferase |
chr16 |
|
CYP2B7P1 |
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1 |
chr19 |
|
SLC10A1 |
solute carrier family 10, member 1 |
chr14 |
|
TDO2 |
tryptophan 2,3-dioxygenase |
chr4 |
|
CYP2A6|CYP2A7|CYP2A13 |
cytochrome P450, family 2, subfamily A, polypeptide 6 | 7 | 13 |
chr19 |
|
HSD17B6 |
hydroxysteroid 17-Beta Dehydrogenase 6 |
chr12 |
|
APOF |
apolipoprotein F |
chr12 |
|
HAL |
histidine ammonia-lyase |
chr12 |
|
C9 |
complement component 9 |
chr5 |
|
FCN3 |
ficolin 3 (Hakata antigen) |
chr1 |
|
SLC22A1 |
solute carrier family 22, member 1 |
chr6 |
|
HSD17B13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
chr4 |
|
F9 |
coagulation factor IX |
chrX |
|
HAO2 |
hydroxyacid oxidase 2 (long chain) |
chr1 |
|
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
chr1 |
|
CRP |
C-reactive protein, pentraxin-related |
chr1 |
|
GLYAT |
glycine-N-acyltransferase |
chr11 |
|
ABCB11 |
ATP-binding cassette, sub-family B, member 11 |
chr2 |
|
NNMT |
nicotinamide N-methyltransferase |
chr11 |
|
SLC1A1 |
solute carrier family 1, member 1 |
chr9 |
II Analysis of gene expression according to risk stratification
Clinically, among the several risk stratification systems proposed for HBL, we used the CHIC classification criteria to classify HBL into three risk categories: standard, intermediate, and high risk [9, 10]. The high-risk group consisted of HBL cases with distant metastasis, and the primary tumors showed significantly upregulated expression of TERC (Telomerase RNA Component),?GFRA3 (GDNF Family Receptor Alpha 3), SPINK1 (Serine Peptidase Inhibitor, Kazal Type 1), DUSP9 (Dual Specificity Phosphatase 9), and histone cluster genes such as HIST1H3B. On the other hand, AFM (Aflanin, a member of albmin gene family), HAO1 (Hydroxyacid Oxidase 1), BAAT1 (Bile Acid-CoA: Amino Acid N-Acyltransferase) and cytochrome P450 family genes (CYP3A43, CYP2C9 etc.) were downregulated. In the comparison the standard- and intermediate-risk groups, SPINK1 and KRT23 (Keratin 23) were upregulated more than twofold, and HSD17B2 (Hydroxysteroid 17-Beta Dehydrogenase 6), PIPOX (Pipecolic Acid And Sarcosine Oxidase), ABCAB (ATP-binding cassette, sub-family B, member 11), CYP8B1, BDH1 (3-Hydroxybutyrate Dehydrogenase 1), and XPNPEP2 (X-Prolyl Aminopeptidase 2) were downregulated more than twofold. These down regulate genes were correlated with hepatocyte differentiation. The pathway analysis of these genes indicated downregulation of bile acid biosynthesis signaling and correlate with liver hyperplasia and hyperproliferation.
Figure 1: Network analysis using the upregulated or downregulated genes in progress ion or relapse cases
The upregulated or downregulated genes more than twofold with the comparison of the relapse or progression cases to the other HBL cases. The genes upregulated or downregulated genes more than twofold are listed in Table 3. Red marks are up-regulated and blue marks are down-regulated genes. In this network, the core upstream genes of this network are HNF4A (Hepatocyte Nuclear Factor 4 Alpha), GSTZ1 (Glutathione S-Transferase Zeta 1), NFE2L3 (Nuclear Factor, Erythroid 2 Like 3) and ESR1 (Estrogen Receptor 1).
III Gene expression analysis in the primary tumors of HBL progression or relapse cases
Among the HBL cases evaluated, 19 were patients with relapse or progression, and 13 were patients who subsequently died of HBL. The gene expression profiles of the relapse or progression cases were compared with those of the other HBL cases. The genes upregulated or downregulated genes more than twofold are listed in (Table 3). Among the upregulated genes, AFP is located on Chr. 4, SPINK1, and PGC (Progastricsin) were on chromosome 5, and NQO1 (NAD(P)H Quinone Dehydrogenase 1) is on chromosome 16. The downregulated genes were GLYAT (Glycine-N-Acyltransferase), located on chromosome 11, FMO3 (flavin containing monooxygenase 3), GYS2 (glycogen synthase 2), BAAT (bile acid Coenzyme A), SLCO1B3 (solute carrier organic anion transporter family, member 1B3), HAO1 (hydroxyacid oxidase (glycolate oxidase) 1), CYP2C19(cytochrome P450, family 2, subfamily C), polypeptide 19), SLC38A4 (solute carrier family 38, member 4), SLC10A1 (solute carrier family 10 (sodium/bile acid cotransporter family), member 1), ABCA6(ATP-binding cassette, member 6), and PON1 (paraoxonase 1). GLYAT located on chromosome 11 and CYP2C19, SLCO1B3, GYS2, and SLC38A4, located on chromosome 12.
III Gene expression analysis in the primary tumors of HBL progression or relapse cases
The pathway analysis of these genes indicated upregulation of FXR /RXR, which is associated with regulation of HNF4A (Figure 1). In this network, the core upstream genes of this network are HNF4A, GSTZ1 (Glutathione S-Transferase Zeta 1), NFE2L3 (Nuclear Factor, Erythroid 2 Like 3) and ESR1 (Estrogen Receptor 1).
IV Clustering analysis
The results of unsupervised clustering of 16 genes up- and downregulated more than twofold is shown in (Figure 2). We found three clusters. In the upper cluster, none of the 20 patients died of HBL, whereas in the other two clusters, 13 of 33 patients died of HBL (P < 0.01). Interestingly, the two oldest patients (7 and 8 years old) with low-risk tumors were included in the upper cluster, whereas most of those represented in the bottom cluster were less than 1 year old. CTNNB1 gene alterations and histological classification were not associated with these clusters, suggesting that these gene expression profiles may be independent prognostic factors for HBL.? Since these patients were treated in the same clinical trial, the gene expression profiles provide information on the biological characteristics of HBL.
Discussion
There are some previous reports of gene expression data in HBL samples, but no data from untreated HBL patients who were enrolled in the clinical trial and treated under the same regimen have been reported [15-18]. This study consisted of tumor samples from patients who participated in the JPLT-2, and all tumor samples were collected prior to treatment. Therefore, the gene expression data of our series were not influenced by treatments such as preoperative chemotherapy and vascular intervention
The up- and down-regulated genes in the comparison between paired NLT and HBL tumor RNA showed activation of Wnt signaling genes and down-regulation of liver function correlating enzyme such as tyrosine aminotransferase and apolioprotein F. The network analysis of the genes differentially expressed between HBL and NLT samples suggested CTNNB1, HNF1A, and HNF4A as upstream transcription factors. The CTNNB1 gene is mutated at a high frequency in HBL, supporting our identification of these genes as distinguishing expressed in HBL. In addition, CYP family members were among the genes downregulated in HBL. These expression profiles suggest that activation of hepatocyte proliferation and inhibition of differentiation are associated with HBL. Among the upregulated genes, DKK1 is an inhibitor of Wnt signaling, and GPC3 overexpression has been reported previously in HBL as well as nephroblastoma [19-22]. Therefore, these gene expression profiles seemed to be reliable, and the oncogenesis of HBL may depend on aberrant Wnt signaling and ontogenesis.
To clarify the biological differences in HBL, we analyzed differences in the gene expression profiles according to risk stratification. Almost all high-risk HBL cases had distant metastasis and high expression of histone cluster genes and small nucleolar RNA, suggesting that distant metastasis of HBL may be correlated with epigenetic regulation. Among these upregulated genes, TERC is the essential component of human telomerase, which is activated in human stem cells and immortalized cells. Our previous study already indicated the high telomerase activity of HBL is correlated with poor outcome of the patients [5, 23]. Therefore, telomerase activation and TERC expression might be the key factor for HBL progression. In the comparison between standard- and intermediate-risk tumors, the downregulation of genes involved in the SIBLING signaling pathway indicated that inhibition of tissue differentiation may be associated with local tumor infiltration. As tissue differentiation is regulated by epigenetic modifications, HBL development and progression may also be related to epigenetic regulation [24, 25]. The gene expression profiles of the HBL progression or relapse cases showed upregulation of four genes: AFP, SPINK1, PGC, and NQO1. An association between AFP overexpression and HBL progression has been reported previously [18]. These data also suggested that epigenetic regulation plays a role in HBL progression or relapse, because the differentially expressed genes are restricted to specific chromosomes. Chromosomal aberrations, including loss of chromosome 11, have been correlated with tumor progression [6, 26]. Some differentially expressed genes may result from chromosomal aberrations, including those in chromosomes 1 and 2, where had already reported as frequently aberrant regions in HBL [27]. Further analyses including genomic aberrations should be performed for clarification of the genomic characteristics of HBL.
Figure 2: Unsupervised clustering of gene expression profiles of hepatoblastoma (HBL) tissue samples
Cluster heatmap of the expression (median centered by row) of 16 genes that were more than two-fold up- or downregulated in all 53 HBL tissue samples. Clustering analysis revealed three distinct clusters: (A) upper, (B) middle, and (C) bottom. Of the 33 cases represented in the upper and middle clusters, 13 subsequently died of HBL, whereas none in the upper cluster (n = 20) died of HBL (P < 0.01).
The clustering analysis of the genes differentially expressed in the relapse or progression cases revealed three major tumor clusters (Figure 2). There were several differences among these HBL cases in terms of histology and patient age at onset. All deceased cases (n = 13) were included in the top and middle clusters, suggesting that this clustering analysis may be useful for risk stratification of HBL tumors. Sixteen genes discriminated in invasive and metastatic HBL samples by Cairo et al. were not included among our genes differentially expressed more than twofold, except for AFP [18]. Since most of the samples in their study were isolated after chemotherapy, the difference in gene expression profiles compared with our study may have been due to treatment effects. Our pathway analysis of the gene set in (Table 3) revealed significant activation of RXR pathway and inhibition of LPS/IL-1 mediated pro-inflammatory cytokine signaling which leads to impaired metabolism, transport and/or biosynthesis of lipid, cholesterol, bile acid and xenobiotics. These genes may provide novel therapeutic targets.
Table 3: List of up- and downregulated genes in tissues from hepatoblastoma recurrence or progression cases
|
|
Fold change |
|
|
Upregulated (> 2-fold) |
||
|
SPINK1 AFP PGC NQO1 |
serine peptidase inhibitor, Kazal type 1 alpha-fetoprotein progastricsin (pepsinogen C) NAD(P)H dehydrogenase, quinone 1 |
2.92124 2.49094 2.45864 2.15604 |
|
Downregulated ( > 2-fold) |
||
|
GLYAT FMO3 GYS2 BAAT SLCO1B3 |ELMOD3 HAO1 CYP2C19 SLC38A4 C7orf58 SLC10A1
ABCA6 PON1 |
glycine-N-acyltransferase flavin containing monooxygenase 3 glycogen synthase 2 bile acid coenzyme A: amino acid N-acyltransferase solute carrier organic anion transporter family, member 1B3 | ELMO/CED-12 domain containing 3 hydroxyacid oxidase (glycolate oxidase) 1 cytochrome P450 family 2 subfamily C member 9 solute carrier family 38 member 4 chromosome 7 open reading frame 58 solute carrier family 10 (sodium/bile acid cotransporter family), member 1 ATP-binding cassette, sub-family A (ABC1), member 6 paraoxonase 1 |
−2.57142 −2.51047 −2.44638 −2.21552 −2.16548
−2.15545 −2.12131 −2.11507 −2.11388 −2.07694
−2.03363 −2.01788 |
Recent developments in whole-genome or exome analyses in tumor biology have revealed genomic alterations in childhood tumors including HBL. Further analysis of genomic alterations in HBL will be necessary to elucidate the driver mutations for diagnosis and targeted therapy in HBL.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This research was partially supported by the Practical Research for Innovative Cancer Control from the Japan Agency for Medical Research (AMED) (NO. JP18CK0106332 and JP18LK0201066) and Grant-in-Aids for Scientific Research (A) (No. 15H02567) from the Ministry of Education, Culture, Sports, and Science of Japan. We would like to thank all staff of the institutes that participated in the JPLT for enrolling their patients in this study. We thank Mrs. S. Hirano, Ms. I. Fukuba, and Mrs. F. Irisuna for the technical assistance. We also thank the Analysis Center of Life Science, Natural Science Center of Basic Research and Development, and the Research Center for Molecular Center, Graduate School of Biomedical Science, Hiroshima University, for the use of their facilities.
Abbreviations
|
AFP |
α-fetoprotein |
|
CHIC |
Childhood Hepatic tumors International Collaboration |
|
COG |
Children’s Oncology Group |
|
HBL |
hepatoblastoma |
|
JPLT |
Japanese Study Group for Pediatric Liver Tumors |
|
NCL |
noncancerous liver tissue |
|
PRETEXT |
PRE-Treatment EXTent of tumor |
|
SIOPEL |
International Childhood Liver Tumor Strategy Group |
Article Info
Article Type
Original ArticlePublication history
Received: Wed 09, Jan 2019Accepted: Fri 25, Jan 2019
Published: Tue 12, Feb 2019
Copyright
© 2023 Eiso Hiyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.EJMC.2018.01.003
Author Info
Masami Kanawa Eiso Hiyama Kasumi Kawashima Keiko Hiyama Kyoko Ikeda Nagisa Morihara Sho Kurihara Takahiro Fukazawa Yuka Ueda
Corresponding Author
Eiso HiyamaNatural Science Center for Basic Research and Development (N-BARD)
Figures & Tables
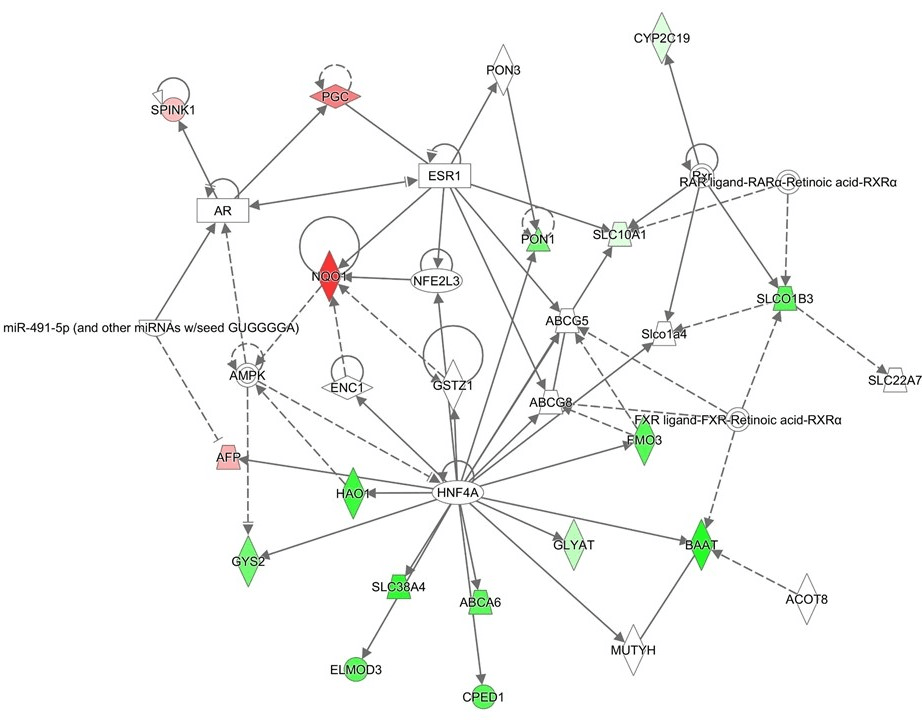

Table 1. Clinicopathological features of the hepatoblastoma samples used for microarray analysis.
|
Age (months) |
|
|
Median |
22 |
|
Range |
0-109 |
|
Sex |
|
|
Female |
25 (47.2%) |
|
Male |
28 (52.8%) |
|
Histologic type |
|
|
Well differentiated |
30 (56.6%) |
|
Poorly differentiated |
21 (39.6%) |
|
Other |
2 (3.8%) |
|
PRETEXT classificationa |
|
|
I |
9 (18.0%) |
|
II |
15 (28.3%) |
|
III |
18 (34.0%) |
|
IV |
11 (20.7%) |
|
Distant metastasis |
14 (26.4%) |
|
CHIC risk stratificationb |
|
|
Standard |
31 (58.5%) |
|
Intermediate |
8 (15.1%) |
|
High |
14 (26.4%) |
|
CTNNBI gene alteration |
|
|
Deletion |
23 (43.4%) |
|
Mutation (exon 3) |
16 (18.9%) |
|
Event |
|
|
Free |
32 (60.4%) |
|
+ |
21 (39.6%) |
|
Clinical course |
|
|
Alive |
38 (71.7%) |
|
Dead |
15 (28.3%) |
aPRETEXT: PRETreatment of EXTent of disease 2,7, bCHIC: Children Hepatic tumor International Collaboration 9,10
Table 2: Up- and downregulated genes in hepatoblastoma tissues compared with noncancerous tissues
|
Gene symbol |
Gene |
Location |
|
Upregulated genes |
||
|
REG3A |
regenerating islet-derived 3 alpha |
chr2 |
|
DPEP1 |
dipeptidase 1 (renal) |
chr16 |
|
SNORD114-26 |
small nucleolar RNA, C/D box 114-26 |
chr14 |
|
LGR5 |
leucine-rich repeat-containing G protein-coupled receptor 5 |
chr12 |
|
DKK1 |
dickkopf homolog 1 (Xenopus laevis) |
chr10 |
|
EPCAM |
epithelial cell adhesion molecule |
chr4 |
|
TSPAN5 |
tetraspanin 5 |
chr4 |
|
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
chr18 |
|
SNORD113-4 |
small nucleolar RNA, C/D box 113-4 |
chr14 |
|
NOTUM |
notum pectinacetylesterase homolog (Drosophila) |
chr17 |
|
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
chr12 |
|
NKD1 |
naked cuticle homolog 1 (Drosophila) |
chr16 |
|
DKK4 |
dickkopf homolog 4 (Xenopus laevis) |
chr8 |
|
MEP1A |
meprin A, alpha (PABA peptid e hydrolase) |
chr6 |
|
SLC7A11 |
solute carrier family 7, member 11 |
chr4 |
|
PEG10 |
paternally expressed 10 |
chr7 |
|
DLK1 |
delta-like 1 homolog (Drosophila) |
chr14 |
|
C9orf4 |
chromosome 9 open reading frame 4 |
chr9 |
|
GPC3 |
glypican 3 |
chrX |
|
Downregulated genes |
||
|
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
chr19 |
|
CYP2C8|CYP2C19 |
cytochrome P450, family 2, subfamily C, polypeptide 8 | 19 |
chr10 |
|
TAT |
tyrosine aminotransferase |
chr16 |
|
CYP2B7P1 |
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1 |
chr19 |
|
SLC10A1 |
solute carrier family 10, member 1 |
chr14 |
|
TDO2 |
tryptophan 2,3-dioxygenase |
chr4 |
|
CYP2A6|CYP2A7|CYP2A13 |
cytochrome P450, family 2, subfamily A, polypeptide 6 | 7 | 13 |
chr19 |
|
HSD17B6 |
hydroxysteroid 17-Beta Dehydrogenase 6 |
chr12 |
|
APOF |
apolipoprotein F |
chr12 |
|
HAL |
histidine ammonia-lyase |
chr12 |
|
C9 |
complement component 9 |
chr5 |
|
FCN3 |
ficolin 3 (Hakata antigen) |
chr1 |
|
SLC22A1 |
solute carrier family 22, member 1 |
chr6 |
|
HSD17B13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
chr4 |
|
F9 |
coagulation factor IX |
chrX |
|
HAO2 |
hydroxyacid oxidase 2 (long chain) |
chr1 |
|
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
chr1 |
|
CRP |
C-reactive protein, pentraxin-related |
chr1 |
|
GLYAT |
glycine-N-acyltransferase |
chr11 |
|
ABCB11 |
ATP-binding cassette, sub-family B, member 11 |
chr2 |
|
NNMT |
nicotinamide N-methyltransferase |
chr11 |
|
SLC1A1 |
solute carrier family 1, member 1 |
chr9 |
Table 3: List of up- and downregulated genes in tissues from hepatoblastoma recurrence or progression cases
|
|
Fold change |
|
|
Upregulated (> 2-fold) |
||
|
SPINK1 AFP PGC NQO1 |
serine peptidase inhibitor, Kazal type 1 alpha-fetoprotein progastricsin (pepsinogen C) NAD(P)H dehydrogenase, quinone 1 |
2.92124 2.49094 2.45864 2.15604 |
|
Downregulated ( > 2-fold) |
||
|
GLYAT FMO3 GYS2 BAAT SLCO1B3 |ELMOD3 HAO1 CYP2C19 SLC38A4 C7orf58 SLC10A1
ABCA6 PON1 |
glycine-N-acyltransferase flavin containing monooxygenase 3 glycogen synthase 2 bile acid coenzyme A: amino acid N-acyltransferase solute carrier organic anion transporter family, member 1B3 | ELMO/CED-12 domain containing 3 hydroxyacid oxidase (glycolate oxidase) 1 cytochrome P450 family 2 subfamily C member 9 solute carrier family 38 member 4 chromosome 7 open reading frame 58 solute carrier family 10 (sodium/bile acid cotransporter family), member 1 ATP-binding cassette, sub-family A (ABC1), member 6 paraoxonase 1 |
−2.57142 −2.51047 −2.44638 −2.21552 −2.16548
−2.15545 −2.12131 −2.11507 −2.11388 −2.07694
−2.03363 −2.01788 |
References
- (2002) Novel therapeutic approaches in the treatment of children with hepatoblastoma. J Pediatr Hematol Oncol 24: 751-755. [Crossref]
- Perilongo G, Shafford E, Plaschkes J (2000) SIOPEL trials using preoperative chemotherapy in hepatoblastoma. Lancet Oncol 1: 94-100. [Crossref]
- Sasaki F, Matsunaga T, Iwafuchi M, Hayashi Y, Ohkawa H, et al. (2002) Outcome of hepatoblastoma treated with the JPLT-1 (Japanese Study Group for Pediatric Liver Tumor) Protocol-1: A report from the Japanese Study Group for Pediatric Liver Tumor. J Pediatr Surg 37: 851-856. [Crossref]
- Hiyama E, Ueda Y, Onitake Y, Kurihara S, Watanabe K, et al. (2013) A cisplatin plus pirarubicin-based JPLT2 chemotherapy for hepatoblastoma: experience and future of the Japanese Study Group for Pediatric Liver Tumor (JPLT). Pediatr Surg Int 29: 1071-1075. [Crossref]
- Hiyama E, Yamaoka H, Matsunaga T, Hayashi Y, Ando H, et al. (2004) High expression of telomerase is an independent prognostic indicator of poor outcome in hepatoblastoma. Br J Cancer 91: 972-979. [Crossref]
- von Schweinitz D, Kraus JA, Albrecht S, Koch A, Fuchs J, et al. (2002) Prognostic impact of molecular genetic alterations in hepatoblastoma. Med Pediatr Oncol 38: 104-108. [Crossref]
- Hishiki T, Matsunaga T, Sasaki F, Yano M, Ida K, et al. (2011) Outcome of hepatoblastomas treated using the Japanese Study Group for Pediatric Liver Tumor (JPLT) protocol-2: report from the JPLT. Pediatr Surg Int 27: 1-8. [Crossref]
- Hata Y (1990) The clinical features and prognosis of hepatoblastoma: follow-up studies done on pediatric tumors enrolled in the Japanese Pediatric Tumor Registry between 1971 and 1980. Part I. Committee of Malignant Tumors, Japanese Society of Pediatric Surgeons. Jpn J Surg 20: 498-502. [Crossref]
- Czauderna P, Haeberle B, Hiyama E, Rangaswami A, Krailo M, et al. (2016) The Children's Hepatic tumors International Collaboration (CHIC): Novel global rare tumor database yields new prognostic factors in hepatoblastoma and becomes a research model. Eur J Cancer 52: 92-101. [Crossref]
- Meyers RL, Maibach R, Hiyama E, Häberle B, Krailo M, et al. (2017) Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children's Hepatic tumors International Collaboration. Lancet Oncol 18: 122-131. [Crossref]
- Haas JE, Muczynski KA, Krailo M, Ablin A, Land V, et al. (1989) Histopathology and prognosis in childhood hepatoblastoma and hepatocarcinoma. Cancer 64: 1082-1095. [Crossref]
- Chromczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol -chloroform extraction. Anal Biochem 162: 156-159.
- Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, et al. (1999) Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res 59: 269-273. [Crossref]
- Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H (2007) RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov 6: 793-810. [Crossref]
- Bhusari S, Pandiri AR, Nagai H, Wang Y, Foley J, et al. (2015) Genomic Profiling Reveals Unique Molecular Alterations in Hepatoblastomas and Adjacent Hepatocellular Carcinomas in B6C3F1 Mice. Toxicol Pathol 43: 1114-1126. [Crossref]
- Kawamoto S, Ohnishi T, Kita H, Chisaka O, Okubo K (1999) Expression profiling by iAFLP: A PCR-based method for genome-wide gene expression profiling. Genome research 9: 1305-1312. [Crossref]
- Lopez-Terrada D (2006) Integrating the diagnosis of childhood malignancies. Adv Exp Med Biol 587: 121-137. [Crossref]
- Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, et al. (2008) Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell 14: 471-484. [Crossref]
- Lopez-Terrada D, Gunaratne PH, Adesina AM, Pulliam J, Hoang DM, et al. (2009) Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol 40: 783-794. [Crossref]
- Midorikawa Y, Ishikawa S, Iwanari H, Imamura T, Sakamoto H, et al. (2003) Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer 103: 455-465. [Crossref]
- Kinoshita Y, Tanaka S, Souzaki R, Miyoshi K, Kohashi K, et al. (2015) Glypican 3 expression in pediatric malignant solid tumors. Eur J Pediatr Surg 25: 138-144. [Crossref]
- Zhou S, O'Gorman MR, Yang F, Andresen K, Wang L (2017) Glypican 3 as a Serum Marker for Hepatoblastoma. Sci Rep 7: 45932.
- Ueda Y, Hiyama E, Kamimatsuse A, Kamei N, Ogura K, et al. (2011) Wnt signaling and telomerase activation of hepatoblastoma: correlation with chemosensitivity and surgical resectability. J Pediatr Surg 46: 2221-2227.[ Crossref]
- Honda S, Minato M, Suzuki H, Fujiyoshi M, Miyagi H, et al. Clinical prognostic value of DNA methylation in hepatoblastoma: Four novel tumor suppressor candidates. Cancer Sci 07: 812-819. [ Crossref]
- Honda S1, Miyagi H, Suzuki H, Minato M, Haruta M, et al. R(2003) ASSF1A methylation indicates a poor prognosis in hepatoblastoma patients. Pediatric surgery international 29: 1147-1152. [Crossref]
- Little MH, Thomson DB, Hayward NK, Smith PJ (1988) Loss of alleles on the short arm of chromosome 11 in a hepatoblastoma from a child with Beckwith-Wiedemann syndrome. Hum Genet 79: 186-189. [ Crossref]
- Mullarkey M, Breen CJ, McDermott M, O'Meara A, Stallings RL (2001) Genetic abnormalities in a pre and post-chemotherapy hepatoblastoma. Cytogenet Cell Genet 95: 9-11. [Crossref]
