Bell’s Palsy May Associate with Increased IL1R1: A Pilot Novel Gene and lncRNA Study
A B S T R A C T
Background: Bell’s palsy is a widespread disease of the peripheral nervous system which causes not only physical disorders but also mental suffering as well. However, the etiological factor of Bell’s palsy is still unclear. The present study aimed to search for potential influencing factors by identifying the key genes and long non-coding RNAs (lncRNAs) involved in patients with Bell’s palsy using RNA-Seq data based on bioinformatics tools.
Methods: Differentially expressed genes (DEGs) and differentially expressed lncRNAs (DELs) in patients before and after therapy, and that in normal control group were identified. The competing endogenous RNAs (ceRNAs) regulatory network was constructed by integrating lncRNA-mRNA pairs, miRNA-mRNA regulatory pairs, and miRNA-lncRNA pairs using Cytoscape. The Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses of DEGs and DELs were evaluated to explore their functions. The targeted corresponding genes and pathogens were verified by ELISA and q-PCR.
Results: In the present study, hub proteins such as CXCR2 and IL1R1 in PPI network and 1039 lncRNA-mRNA co-expression pairs (eg., CYYR1-AS1-MDM2) were identified. 739 miRNA-mRNA pairs (eg., hsa-miR-147a-IL1R1), 255 miRNA-lncRNA pairs, and 363 mRNA-lncRNA co-expression pairs were included in ceRNA regulatory network. Meanwhile, CYYR1-AS1 was enriched into most pathways, including Epstein-Barr virus (EBV) infection. Subsequently, validation of neuroinflammation relevant IL1R1 and EBV showed that IL1R1 was upregulated in the serum of patients with Bell’s palsy before therapy, while EBV was not found among them.
Conclusion: We hypothesized that etiological factor of Bell's palsy correlate to complex miRNA-lncRNA-mRNA interacting networks and IL1 might be involved in inflammation and immune regulation in the onset of Bell's palsy.
Keywords
Bell’s palsy, microRNAs, long non-coding RNA, competing endogenous RNAs, IL1R1, Epstein-Barr virus
Introduction
Bell’s palsy (BP) is a type of peripheral facial paralysis, which leads to the weak or paralyzed facial muscles on the affected side [1]. Epidemiological report suggests a high annual incidence of 23-37 per 100,000 per year in UK, and most commonly affects people of 15-45 years old [2, 3]. Diabetes and viral infection are determined as risk factors for Bell's palsy development [4, 5]. So far, anti-inflammatory or antivirus drugs such as corticosteroids and aciclovir have been used to improve outcomes [6]. However, development of further effective treatment has been limited due to unclear pathogenesis of the disease. This study aimed to search for potential influencing factors by identifying the key genes and long non-coding RNAs (lncRNAs) and their relevant networks involved in patients with Bell’s palsy using RNA-Seq data based on bioinformatics tools, which will be helpful to reveal the underlying mechanism of the onset of BP.
Methods
I RNA Extraction and Illumina Sequencing
Serum samples of total 15 patients with Bell’s palsy were collected at the timepoint of disease onset (before therapy group) and complete recovery (after therapy group) were collected in this study. And another 15 normal controls were set as normal control group. The total RNA from each sample was extracted using Trizol reagent (Invitrogen, 15596-018, USA). After the quality control of RNA, mRNA was enriched by oligo (dT) magnetic beads, and then broke into shot fragments by fragmentation buffer. Then, the RNA fragments were reverse transcribed into the first strand cDNA, and the second strand cDNA was compounded. The final cDNA library was constructed after double strands cDNA were purified and repaired. Then, the cDNA libraries were sequenced on an Illumina HiSeq™ 3500. The raw sequencing data have been uploaded to the public database NCBI. The reads with N content exceeding 10% of total base number and more than 50% low quality base of total base number (Q≤5) were deleted.
II Data Comparison and Transcriptional Annotation
Following sample sequencing and data filtration, the obtained clean reads were matched to the human reference genome derived from GENCODE (Release 25, GRCh38) using Tophat (v2.1.0) software [7]. Then, based on the annotation information, the read counts of each matched gene were obtained using featureCounts software (v1.6.0) [8].
III Identification of DEGs and DELs
The DEGs and DELs between after therapy group vs before therapy group, before therapy group vs normal control group, and after therapy group vs normal control group were identified using the edgeR package (version 3.4, (Link 1)), respectively [9, 10]. Thresholds of DEGs and DELs were defined as Log fold-change (FC)> 0.585 and P value < 0.05. Then, the overlapping DEGs and DELs with opposite expression trend in the two comparisons (after therapy vs before therapy and before therapy vs normal control) were selected. After removing the DEGs and DELs identified between after therapy group and normal control groups from above genes, the remaining shared DEGs and DELs were screened for further analyses. Subsequently, heatmaps of these DEGs and DELs in three groups were drawn using pheatmap (version 1.0.8, (Link 2)) in R software.
IV GO and KEGG Pathway Analyses for DEGs
The GO-biological process terms and KEGG pathway of DEGs were enriched by clusterProfiler package (version 3.8.1) in R software [11]. Benjamini and Hochberg (BH) method was utilized to adjust the enrichment statistic P values. The significant GO-biological process terms and KEGG pathways for DEGs were selected with cut off of P. adjust < 0.05.
V Constructing Protein-Protein Interaction (PPI) Network Based on DEGs
STRING (version: 10.0, (Link 3)) was used to analyse PPI between DEGs, and PPI with score (medium confidence) > 0.4 was used to construct network [12]. Then PPI network was visualized by Cytoscape software (Link 4). Subsequently, CytoNCA plug-in (version 2.1.6, (Link 4)) was applied for topological analysis of nodes in PPI network [13]. Parameter was set as ‘without weight’, and the result of Degree Centrality (DC) was output. At last, the hub protein was identified according to their DC ranking.
VI Co-Expression Analysis Between DEGs and DELs
Firstly, Pearson’s correlation coefficient of each DEG and DEL was calculated. Then, thresholds of P<0.05 and correlation coefficient (r)>0.8 were used to identify significantly co-expressed lncRNA-mRNA pairs.
VII Function and Pathway Analyses for DELs
The function prediction of DELs was analysed using its corresponding DEGs in above screened significantly co-expressed lncRNA-mRNA pairs. Similarly, the GO and KEGG pathway terms of corresponding DEGs were enriched by clusterProfiler package (version 3.8.1). The enrichment statistic P values were adjusted by BH method, and significant results were obtained under P<0.05.
VIII Identification of miRNA-lncRNA and miRNA-mRNA Interaction Pairs
The miRNAs relative to lncRNA and mRNA in above screened co-expressed lncRNA-mRNA pairs were analysed, respectively. miRNA-lncRNA pairs were predicted using lnCeDB database based on miranda tool. In addition, miRWALK2.0 (Link 5) database, which integrated the information of 12 databases (including miRWalk, Microt4, miRanda, mirbridge, miRDB, miRMap, miRNAMap, Pictar2, PITA, RNA22, RNAhybrid and Targetscan), was used to identify upstream miRNA of mRNA (species: Homo). Finally, shared miRNA-mRNA interactions in at least 7 above databases were used.
IX Comprehensive Analysis of ceRNA Regulatory Network
ceRNA regulatory network was constructed by integrating the lncRNA-miRNA interaction pairs, miRNA-mRNA pairs, and lncRNA-mRNA co-expressed pairs (r>0.8) using Cytoscape. Subsequently, the degree of nodes in ceRNA network was analysed using CytoNCA plugin in Cytoscape. Parameter was set as without weight, and nodes with the higher degree were more important in the network.
X IL1R1 and EBV Specific IgG Antibody Detections by ELISA
Serum of 45 BP patients and 32 normal individuals were tested for the presence of IL1R1 and EBV using IL1R1 ELISA kit (eBioscience, USA) and Epstein-Barr Virus-Viral Capsid Antigen (EBV-VCA) IgM, Human, BioAssay ELISA Kit (USBiological, Beijing, China) according to the manufacturer’s instruction. Cut-off was defined with positive and negative control serum that were included in each assay, according to the manufacturer's instruction.
XI Quantification of Plasmatic EBV DNA and IL1R1 RNA
Plasma from 45 BP patients and 32 normal individuals were recovered and conserved at -80°C until use. A volume of 200 µl of plasma was used to extract DNA using QIAamp DNA Mini Kit (Qiagen, France). Circulating levels of EBV DNA were measured by real-time quantitative polymerase chain reaction (q-PCR). The q-PCR reaction was performed using the TaqMan PCR Kit-24 Reactions (Norgenbiotek, CA) on ABI Prism 7700 sequence detection system (Applied Biosystems). The thermal cycling conditions were 95°C for 10 min and 45 cycles of 95°C for 15 sec, 58°C for 1 min, 68°C for 30 sec. For each run, two negative controls without any DNA template were included. Primers of PCR for detection of EBV DNA are Forward: 5′-TGGAAAC CCGTCA CTCTC-3′, Reverse: 5′-TA-ATGGCATAGGTGGAATG-3′.
Total RNA was extracted using the Ultraspec Phenol Kit (Biotecx, USA) according to the manufacturer’s instructions. Then, cDNA was synthesized from total RNA using the cDNA Synthesis Kit (Roche, Germany) and TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, USA). qRT-PCR detected the levels of IL1R1 using an SYBR-green detection system on an ABI-7500 Real-time PCR System (Applied Biosystems, USA). The mRNA expression was expressed relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control, while miRNA expression was expressed relative to U6 as an internal control. The relative expression levels of miRNAs were evaluated using the 2-△△ct method and expression levels were normalized relative to those of U6. The PCR amplification reaction was carried out in a 20-μl system, including 1 μl cDNA, as well as 1 μl forward primer and 1 μl reverse primer. The primer sequences used were listed as followed: IL1R1(261 bps): Primer F 5' CTGTCACCAGCCACTAAG 3'; Primer R 5' TTCCCAAGCCCTCTACTC 3'; GAPDH (218bps): Primer F 5' AATCCCATCACCATCTTC 3', Primer R 5' AGGCTGTTGTCATACTTC 3'. All PCR assays were performed in triplicate.
Results
I Identification of DEGs and DELs
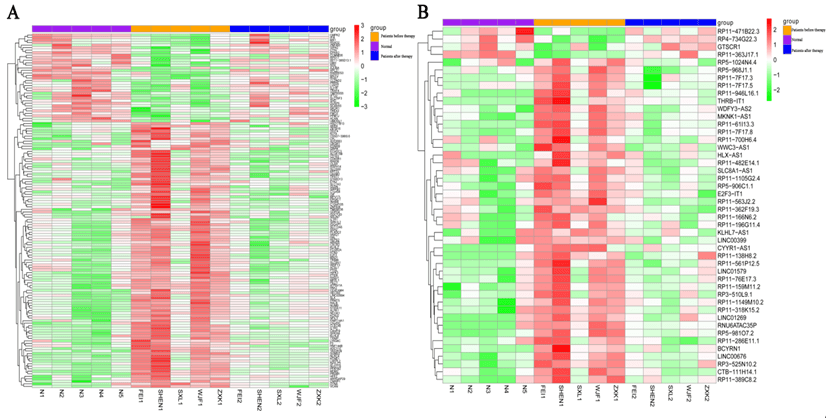
In total, 587, 8339, and 8757 DEGs were identified between the comparison groups of after therapy vs before therapy, before therapy vs normal control, and after therapy vs normal control, respectively. In addition, a total of 214, 3252, and 3319 DELs were identified in the above comparisons (Table 1). Further, a set of 71 shared DELs and 200 shared DEGs with opposite expression trend in the comparisons of after therapy group vs before therapy and after therapy group vs control was obtained. After removing the DEGs and DELs identified between after therapy group and control groups from above genes, a total of 140 DEGs and 46 DELs related to treatment were finally identified. As shown in (Figure 1), the expression patterns of DEGs and DELs in after therapy group were basically consistent with control group, while were opposite with disease group, indicating that medicinal therapy intervention could obviously alter the expressions of mRNA and lncRNA.
Figure 1: Heatmap of A) DEGs and B) DELs. Purple in the top indicates normal control samples; orange in the top indicates Bell’s palsy samples without treatment; dark blue in the top indicates Bell’s palsy samples with treatment. Green indicates down-regulated DEGs or DELs; Red indicates up-regulated DEGs or DELs.
DEGs: Differentially Expressed Genes; DELs: Differentially Expressed lncRNAs.
Table 1: The statistics of differently expressed genes and
lncRNA identified by different group comparisons.
|
|
|
before
therapy vs normal control |
after
therapy vs before therapy |
after
therapy vs normal control |
|
lncRNA |
up |
2424 |
56 |
2454 |
|
down |
828 |
158 |
865 |
|
|
total |
3252 |
214 |
3319 |
|
|
mRNA |
up |
4665 |
125 |
4799 |
|
down |
3674 |
462 |
3958 |
|
|
total |
8339 |
587 |
8757 |
II Functional Analysis of DEGs
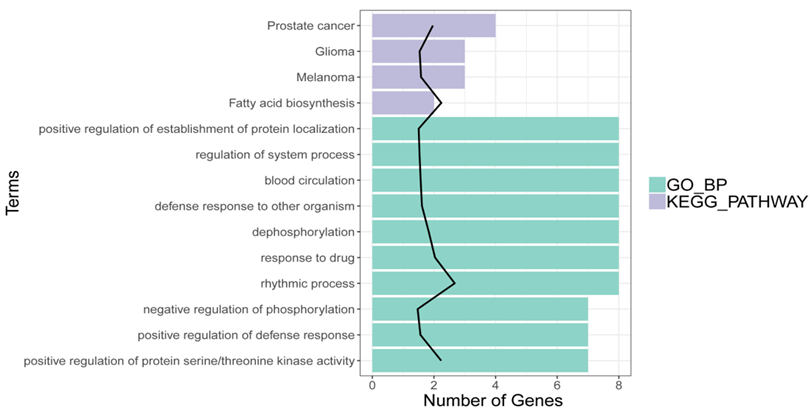
Using pre-set threshold value, the DEGs were totally enriched in 111 GO BP and 4 pathways (Fatty acid biosynthesis, Prostate cancer, Melanoma and Glioma). Among the 111 GO BP terms, the DEGs were closely associated with the terms of “blood circulation” (e.g., CXCR2, MDM and SLC8A1), “negative regulation of phosphorylation”, and “dephosphorylation” were closely associated with (Figure 2).
Figure 2: Results of GO function and KEGG pathway for DEGs. X-axis indicates the count of genes in the function terms and pathways; Y-axis indicates the GO terms and pathways.
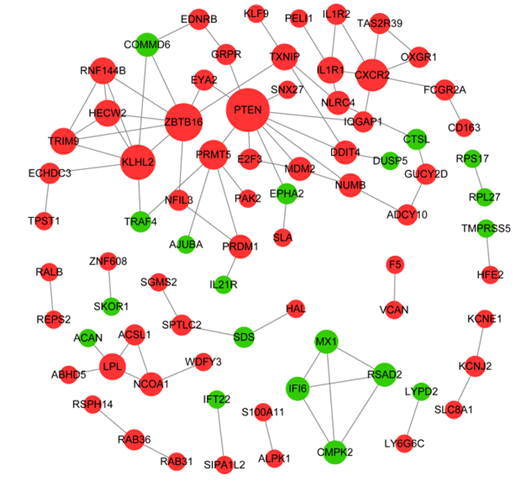
III PPI Network Analysis
In PPI network, there were 78 nodes and 85 interaction pairs, including 59 up-regulated DEGs and 19 down-regulated DEGs (Figure 3). Based on the topological properties analysis, the top 10 hub genes were PTEN (degree=10), ZBTB16 (degree=8), KLHL2 (degree=7), CXCR2 (degree=6) and PRMT5 (degree=5), IL1R1 (degree=4), TXNIP (degree=4), TRIM9 (degree=4), and LPL (degree=4), RNF144B (degree=4), respectively.
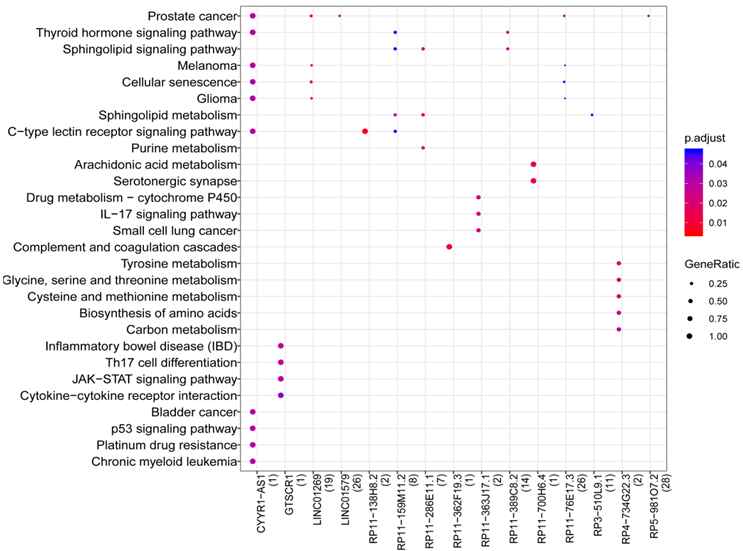
IV lncRNA-mRNA Co-Expression Pairs and lncRNA Functional Prediction
Overall, 1039 lncRNA-mRNA pairs (e.g.) were identified after the lncRNA-mRNA co-expression analysis, including 105 DEGs and 43 DELs. In addition, 15 lncRNAs were enriched to 59 pathways such as Cytokine-cytokine receptor interaction (hsa04060), Sphingolipid metabolism (hsa00600), Arachidonic acid metabolism (hsa00590), and Th17 cell differentiation (hsa04659) (Figure 4). In addition, CYYR1-AS1(cysteine and tyrosine rich 1 antisense RNA 1) was enriched into most pathways, including Epstein-Barr virus infection (MDM2).
V miRNA Prediction and ceRNA Regulatory Network
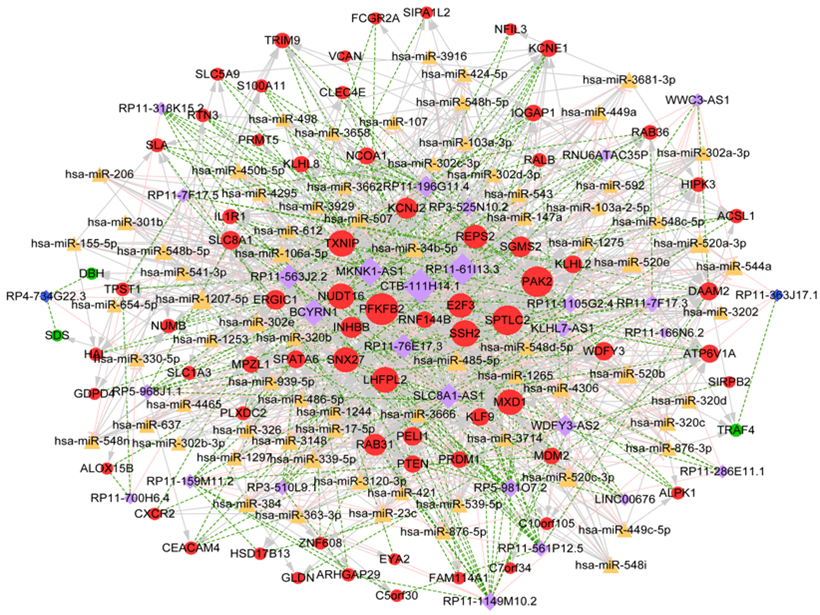
Totally, 3107 miRNA-lncRNA interaction pairs were identified after InCeDB (Link 6), including 844 miRNAs and 35 lncRNAs. Additionally, 10698 miRNA-mRNA interactions were obtained, including 1487 miRNAs and 122 DEGs. Then, the miRNAs (degree ≥ 10) that had both regulatory pairs with co-expressed lncRNA and mRNA were selected, and corresponding ceRNA regulatory network was comprised of 1257 interaction pairs (involving 739 miRNA-mRNA pairs, 255 lncRNA-miRNA pairs and 363 lncRNA-mRNA co-expression pairs) and 174 nodes (involving 29 lncRNA, 73 miRNA and 72 mRNA) (Figure 5). In ceRNA regulatory network, CTB-111H14.1, RP11-61I13.3, PFKFB2, SPTLC2 and PAK2 were the top 5 nodes with higher degree values.
Figure 3: Protein-protein interaction network. Nodes in red circle indicate up-regulated DEGs, while node in green circle indicates down-regulated DEGs. Node size is proportional to its degree value.
Figure 4: Results of pathway prediction for LncRNA. The color changes from red to blue represent the p values from small to large, while the bubble size indicates the proportion of gene counts involved in one term to all genes.
Figure 5: ceRNA regulatory network. Yellow triangle indicates miRNA; light purple rhombus indicates up-regulated lncRNA; dark blue rhombus indicates down-regulated lncRNA red circle indicates up-regulated genes, green circle indicates down-regulated genes. The higher the degree value, the more important the node is.
VI Validation of IL1R1 and EBV
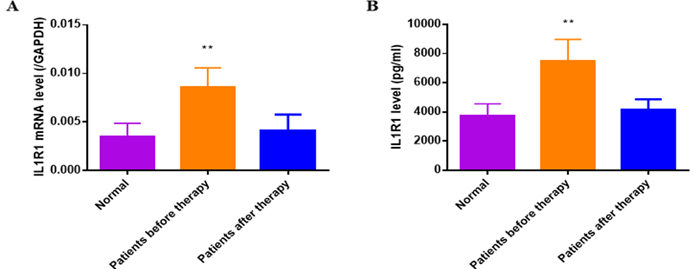
Serum IL1R1 of patients with Bell's palsy and health individuals as control were detected by ELISA and q-PCR analysis. As showed in (Figure 6), IL1R1 was upregulated in patients with Bell's palsy before therapy compared to that in Bell's palsy before therapy(P<0.01) or health control (P<0.01). While serum EBV was not found in any group by ELISA or q-PCR analysis.
Figure 6: The IL1R1 mRNA and protein expression had been analysed by q-PCR and ELISA. A) The serum IL1R1 mRNA expression was up-regulated at the onset of BP compared to that of normal controls(P<0.01), which decreased when the disease completely recovered (P<0.01). B) The serum IL1R1 protein expression of BP patient before therapy was detected higher than normal controls (P<0.01) and that of after therapy (P<0.01).
Discussion
In this study, a total of 140 DEGs and 46 DELs related to medicinal treatment were identified and several hub proteins such as CXCR2 (C-X-C Motif Chemokine Receptor 2) and IL1R1 (Interleukin 1 Receptor Type 1) were identified in PPI network. Subsequently, 1039 lncRNA-mRNA co-expression pairs (eg., CYYR1-AS1-MDM2), 3107 miRNA-lncRNA interaction pairs, and 10698 miRNA-mRNA interactions were obtained. Finally, 739 miRNA-mRNA pairs (eg., hsa-miR-147a-IL1R1), 255 miRNA-lncRNA pairs, and 363 mRNA-lncRNA co-expression pairs were included in ceRNA regulatory network. In addition, among DELs, CYYR1-AS1 was enriched into most pathways, including Epstein-Barr virus infection.
CXCR2, (Link 6) an interleukin 8 (IL-8) receptor, plays a crucial role in neutrophil-driven inflammation and immune functions, motor-neuron degeneration [14, 15]. But CXCR2/IL-8 is more closely associated with tumor-related diseases, such as oesophageal squamous cell carcinoma, colon cancer, and mammary carcinoma, so it was not further verified and discussed in this study [16-19]. But interestingly, another cytokine had come into view which is IL1R1. IL1R1 is a receptor for interleukin-1 (IL-1) alpha and IL-1 beta, which has key roles in many cytokines induced immune and inflammatory responses [20]. A previous study confirms that IL-1, IL-6, and TNF-α levels are increased in serum samples from patients with Bell’s palsy compared with that of healthy populations. We predicted that miR-147a was an upstream regulator of IL1R1. It has been reported that miR-147a as an anti-inflammatory factor, is upregulated by IL-1b stimulation in mesenchymal stem cells [21]. Similarly, upregulation of miR147a and miR147b may decrease the expression of pro-inflammatory factors IL-6 and COX-2 after IL-1β stimulation in astrocyte cells [22]. In this study, IL1R1 was detected highly expressed in patients with Bell's palsy which indicated that IL1 might be involved in inflammation and immune regulation in the onset of Bell's palsy.
Actually, virus can indeed invade the central nervous system and cause paralysis of the facial nerve, including some cases who were diagnosed Bell's palsy that lack rigorous etiology tests. Among them, the most common viruses are the varicella-zoster virus, human herpesvirus 6, herpes simplex virus, enterovirus, and Epstein-Barr virus. In some opinions, virus infection is considered as one main etiology of peripheral facial paralysis [23-25]. Reportedly, Epstein-Barr virus infection is associated with bilateral facial nerve palsy, or even COVID-19 co-infection, including adults and infants [26, 27]. Additionally, in the study of Ramos et al., it shows 70.5% Bell’s palsy cases are positive to Epstein-Barr virus antibodies [19]. It has revealed that viruses may lead to axon degeneration via regulating abnormal expressions of p53 to upregulate apoptosis modulator [28-30].
A possible explanation of Epstein-Barr virus mediated neural dysfunction is the activation of apoptotic pathways and intra-axonal degradation. For example, IL-1 and IL1R were found increased after EBV infection or even in nasopharyngeal carcinoma cell growth [31-33]. IL-1 signaling plays an important role in inflammation and early activation of host innate immune responses following virus infection [34]. But it was both expected and unexpected that no Epstein-Barr virus was found in serum from neither Bell's palsy patients nor normal controls. Here we set a hypothesis that EBV invades the human body under specific conditions, it leads to a significant increase of IL-1 and the damage of facial nerve function. However, EBV might colonize at specific sites but not in peripheral blood, such as facial ganglion, so it had not been detected in serum. Of course, this must be verified by further scientific experiments.
On the other hand, it was predicted in this study that CYYR1-AS1 was enriched in “Epstein-Barr virus infection”, and MDM2 was a co-expressed target of “CYYR1-AS1”. It has been suggested that Epstein-Barr virus nuclear antigen 3C enhances p53 ubiquitination and degradation via deubiquitinating MDM2 [35]. Cysteine and tyrosine-rich 1 (CYYR1)-AS1, located on human chromosome 21, play as a lncRNA to regulate the CYYR1 locus [36]. Although no direct studies have reported a connection of CYYR1-AS1 and Epstein-Barr virus infection, study has suggested that CYYR1 is upregulated in patients with influenza under febrile convulsion, indicating the abnormal expression of CYYR1 or CYYR1-AS1 may be response to virus infection [37].
However, there were some limitations in this study at present. Serum samples rather than tissue samples were detected in this study, so it can only indirectly explain the situation of pathogen infection of nerve tissue, but to harvest a tissue sample in a living individual is really a very difficult task. And the key DELs and DEGs expression in Bell’s palsy patients had not been verified through experiment due to a longtime span in numbers of cases. Based on the current exploratory research with insufficient samples meeting this need, the experiment was not performed in this study. In the future, large scale investigations for the causes of the disease are in need and further confirmation is needed to determine whether there is a viable EB virus in the ganglion. Last but not least, how does Epstein Barr virus invade facial nerve and increase IL1R1 should be put into consideration.
Conclusion
In conclusion, the occurrence and development of Bell facial paralysis is quite complex and may involve related to complex miRNA-lncRNA-mRNA interacting networks and IL1 might be involved in inflammation and immune regulation in the onset of Bell’s palsy. Results of the present study indicated the direction for further exploration in the future.
Ethical Approval
The Institutional Ethics Committee, Baoshan Hospital of Integrated Traditional Chinese Medicine and Western Medicine, Shanghai approved the study (Approval No. 201809-03). This study has been registered at the Chinese Clinical Trials Registry (ChiCTR1800018972).
Consent
All participants had signed informed consent and agreed to supply their blood samples for research use and publication.
Data Availability
The data and material in this study is available from the corresponding author on reasonable request. (Link 8)
Competing Interests
None.
Funding
This study was supported by Shanghai Municipal Health and Family Planning Commission (grant number 201840303); Shanghai Talent Development Fund (grant number 2020086); Key discipline construction found of Baoshan Hospital of Integrated Traditional Chinese Medicine and Western Medicine, Shanghai (grant number BSYYZDZK-2019-03, BSYYZDZK-2019-04).
Author Contributions
Zhidan Liu: Conception, Design and Administrative Support; Xiaoyan Li: Provision of Study Materials or Patients; Chuang Zhao and Chunlan Chen: Collection and Assembly of Data; Xiaoyan Li, Ying Zhao, Chuang Zhao, Zunyuan Li, Wenge Huo: Data analysis and Interpretation; All authors: Manuscript Writing; All authors: Final Approval of Manuscript.
Acknowledgement
Not applicable.
Abbreviation
DEGs: Differentially Expressed Genes
DELs: Differentially Expressed lncRNAs
ceRNAs: Competing Endogenous RNAs
GO: Gene Ontology
KEGG: Kyoto Encyclopedia of Genes and Genomes
EBV: Epstein-Barr Virus
PPI: Protein-Protein Interaction
q-PCR: quantitative Polymerase Chain Reaction
GAPDH: Glyceraldehyde-3-Phosphate Dehydrogenase
CYYR1-AS1: Cysteine and Tyrosine Rich 1 Antisense RNA 1
MDM2: Murine Double Minute2
CXCR2: C-X-C Motif Chemokine Receptor 2
IL1R1: Interleukin 1 Receptor Type 1
Article Info
Article Type
Research ArticlePublication history
Received: Sat 15, Jan 2022Accepted: Sat 29, Jan 2022
Published: Fri 11, Feb 2022
Copyright
© 2023 Zhidan Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.NNB.2022.01.01
Author Info
Zhidan Liu Xiaoyan Li Ying Zhao Chuang Zhao Chunlan Chen Zunyuan Li Wenge Huo
Corresponding Author
Zhidan LiuDepartment of Acupuncture, Baoshan Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Figures & Tables
Table 1: The statistics of differently expressed genes and
lncRNA identified by different group comparisons.
|
|
|
before
therapy vs normal control |
after
therapy vs before therapy |
after
therapy vs normal control |
|
lncRNA |
up |
2424 |
56 |
2454 |
|
down |
828 |
158 |
865 |
|
|
total |
3252 |
214 |
3319 |
|
|
mRNA |
up |
4665 |
125 |
4799 |
|
down |
3674 |
462 |
3958 |
|
|
total |
8339 |
587 |
8757 |
DEGs: Differentially Expressed Genes; DELs: Differentially Expressed lncRNAs.
References
1. Somasundara D,
Sullivan F (2017) Management of Bell’s
palsy. Aust Prescr 40: 94-97. [Crossref]
2.
Morales DR, Donnan PT, Daly F, Staa TV, Sullivan FM
(2013) Impact of clinical trial findings on Bell's palsy management in general
practice in the UK 2001-2012: interrupted time series regression analysis. BMJ Open 3: e003121. [Crossref]
3.
Gagyor I, Madhok VB, Daly F, Sullivan F (2019)
Antiviral treatment for Bell's palsy (idiopathic facial paralysis). Cochrane Database Syst Rev 9: CD001869. [Crossref]
4.
Brandenburg NA, Annegers JF (1993) Incidence and
risk factors for Bell’s palsy in Laredo, Texas: 1974–1982. Neuroepidemiology
12: 313-325. [Crossref]
5.
Morales FS, Illingworth C, Lin K, Agosto IR, Powell
C et al. (2017) PML-IRIS in an HIV-2-infected patient presenting as Bell’s
palsy. J Neurovirol 23: 789-792.
[Crossref]
6.
Khedr EM, Badry R, Ali AM, El Fetoh NA, El Hammady
DH et al. (2016) Steroid/Antiviral for the treatment of Bell’s palsy: Double
blind randomized clinical trial. Restor Neurol Neurosci 34: 897-905. [Crossref]
7.
Ghosh S, Chan CKK (2016) Analysis of RNA-Seq Data
Using TopHat and Cufflinks. Methods Mol Biol 1374: 339-361. [Crossref]
8.
Liao Y, Smyth GK, Shi W (2014) featureCounts: an
efficient general purpose program for assigning
sequence reads to genomic features. Bioinformatics 30: 923-930. [Crossref]
9.
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a
Bioconductor package for differential expression analysis of digital gene
expression data. Bioinformatics 26: 139-140. [Crossref]
10. McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of
multifactor RNA-Seq experiments with respect to biological variation. Nucleic
Acids Res 40: 4288-4297. [Crossref]
11. Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for
comparing biological themes among gene clusters. OMICS 16: 284-287. [Crossref]
12. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D et al. (2015)
STRING v10: protein–protein interaction networks, integrated over the tree of
life. Nucleic Acids Res 43: D447-D452. [Crossref]
13. Tang Y, Li M, Wang J, Pan Y, Wu FX (2015) CytoNCA: a cytoscape plugin for
centrality analysis and evaluation of biological networks. BioSystems 127: 67-72. [Crossref]
14. Jurcevic S, Humfrey C, Uddin M, Warrington S, Larsson B et al. (2015) The
effect of a selective CXCR2 antagonist (AZD5069) on human blood neutrophil
count and innate immune functions. Br J Clin Pharmacol 80: 1324-1336. [Crossref]
15. Ramesh G, MacLean AG, Philipp MT (2013) Cytokines and chemokines at the
crossroads of neuroinflammation, neurodegeneration, and neuropathic pain. Mediators
Inflamm 2013: 480739. [Crossref]
16. David JM, Dominguez
C, Hamilton DH, Palena C (2016) The IL-8/IL-8R Axis: A Double Agent in Tumor
Immune Resistance. Vaccines (Basel) 4: 22. [Crossref]
17. Inoue M, Takeuchi
H, Matsuda S, Nishi T, Fukuda K et al. (2021) IL-8/CXCR2 Signalling Promotes
Cell Proliferation in Oesophageal Squamous Cell Carcinoma and Correlates With
Poor Prognosis. Anticancer Res 41: 783-794. [Crossref]
18. Ma X, Chen J, Liu J, Xu B, Liang X et al. (2021)
IL-8/CXCR2 mediates tropism of human bone marrow-derived mesenchymal stem cells
toward CD133+ /CD44+ Colon cancer stem cells. J Cell
Physiol 236: 3114-3128. [Crossref]
19. Maeda S, Tsuda H, Haruki S, Mitsuto I (1999) Atypical Epstein-Barr virus
infection associated with Gianotti-Crosti syndrome and Bell's palsy. Pediatr Int 41: 315-317. [Crossref]
20. Dinarello CA (2018) Introduction to the interleukin‐1 family of
cytokines and receptors: Drivers of innate inflammation and acquired immunity. Immunol
Rev 281: 5-7. [Crossref]
21. Song Y, Dou H, Li X, Zhao X, Li Y et al. (2017) Exosomal miR‐146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin‐1β‐Primed Mesenchymal Stem Cells
Against Sepsis. Stem Cells 35: 1208-1221. [Crossref]
22. van Scheppingen J, Mills JD, Zimmer TS, Broekaart DWM, Iori V et al.
(2018) miR147b: A novel key regulator of interleukin 1 beta-mediated
inflammation in human astrocytes. Glia 66: 1082-1097. [Crossref]
23. Papan C, Kremp L, Weiß C, Petzold A, Schroten H et al. (2019) Infectious
causes of peripheral facial nerve palsy in children-a retrospective cohort
study with long-term follow-up. Eur J Clin Microbiol Infect Dis 38:
2177-2184. [Crossref]
24.
Vogelnik
K, Matos A (2017) Facial nerve palsy secondary to Epstein-Barr virus infection
of the middle ear in pediatric population may be more common than we think. Wien
Klin Wochenschr 129: 844-847. [Crossref]
25.
Álvarez Argüelles ME,
Rojo Alba S, Rodríguez Pérez M, Abreu Salinas F, de Lucio Delgado A et al. (2019) Infant
Facial Paralysis Associated with Epstein-Barr Virus Infection. Am J Case Rep
20: 1216-1219. [Crossref]
26. Terada K, Niizuma T, Kosaka Y, Inoue M, Ogita S
et al. (2004) Bilateral facial nerve palsy associated with
Epstein-Barr virus infection with a review of the literature. Scand J Infect
Dis 36: 75-77. [Crossref]
27. Cabrera Muras A, Carmona Abellán MM, Collía Fernández A, Uterga Valiente
JM, Antón Méndez L et al. (2021) Bilateral facial nerve palsy associated with
COVID-19 and Epstein-Barr virus co-infection. Eur J Neurol 28: 358-360.
[Crossref]
28. Papaianni E, El Maadidi S, Schejtman A, Neumann S, Maurer U et al. (2015)
Phylogenetically Distant Viruses Use the Same BH3-Only Protein Puma to Trigger
Bax/Bak-Dependent Apoptosis of Infected Mouse and Human Cells. Plos One
10: e0126645. [Crossref]
29. Zhang W, Xu L, Luo T, Wu F, Zhao B et al. (2020) The etiology of Bell’s palsy: a review. J Neurol 267:
1896-1905. [Crossref]
30.
Roberge
CJ, Larochelle B, Rola Pleszczynski M, Gosselin J (1997) Epstein-Barr virus
induces GM-CSF synthesis by monocytes: effect on EBV-induced IL-1 and IL-1
receptor antagonist production in neutrophils. Virology 238: 344-352. [Crossref]
31.
Jabs
WJ, Wagner HJ, Schlenke P, Kirchner H (2000) The primary and memory immune
response to Epstein-Barr virus infection in vitro is characterized by a
divergent production of IL-1beta/IL-6 and IL-10. Scand J Immunol 52:
304-308. [Crossref]
32.
Sabeti
M, Kermani V, Sabeti S, Simon JH (2012) Significance of human cytomegalovirus
and Epstein-Barr virus in inducing cytokine expression in periapical lesions. J
Endod 38: 47-50. [Crossref]
33.
Huang
YT, Liu MY, Tsai CH, Yeh TH (2010) Upregulation of interleukin-1 by
Epstein-Barr virus latent membrane protein 1 and its possible role in
nasopharyngeal carcinoma cell growth. Head Neck 32: 869-876. [Crossref]
34.
Skinner
CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL (2017) An Epstein-Barr Virus
MicroRNA Blocks Interleukin-1 (IL-1) Signaling by Targeting IL-1 Receptor 1. J
Virol 91: e00530-e00517. [Crossref]
35. Saha A, Murakami M, Kumar P, Bajaj B, Sims K et al. (2009) Epstein-Barr
virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and
degradation by deubiquitinating Mdm2. J Virol 83: 4652-4669. [Crossref]
36. Casadei R, Pelleri MC, Vitale L, Facchin F, Canaider S et al. (2014) Characterization of human gene locus CYYR1: a complex multi-transcript system. Mol Biol Rep 41: 6025-6038. [Crossref]
37. Kawada JI, Kimura H, Kamachi Y, Nishikawa K, Taniguchi M et al. (2006) Analysis of gene-expression profiles by oligonucleotide microarray in children with influenza. J Gen Virol 87: 1677-1683. [Crossref]
