Journals
Antisense DNA oligomer targeting of histone deacetylase 1 (HDAC1) mRNA for potential knockdown effects
A B S T R A C T
Histone Deacetylase(HDAC) is an enzyme that eliminates acetyl group from the histone octamer complex. The acetylation state of histone proteins is a major interest of epigenetic gene expression. HDAC1 inhibitors are used for anticancer therapeutics by controlling multiple signaling protein kinases like SAPK, ERK and TNF-alpha. Here, we used single strand DNA 18-oligomer to mimic RNA interference technology. We modeled the HDAC1 mRNA secondary structure and identified the possible four siRNA binding sites by higher possibility than miR-449 targeting site. Also, its possible configuration was modeled according to binding energies. Three places, where the possibility of siRNA binding is low, were randomly identified as positive controls. As a Result, the 18-omer single strand DNA was generated according to the identified sequences. This preliminary data can be further warranted to generate HDAC1 knockdown activity and its comparison to the current HDAC1 inhibitors. Furthermore, generation of single strand DNA as a antisense sequence to a specific mRNA can be utilized for therapeutics along with RNAi. With the thermodynamically stable structure of DNA compared to RNA, it can be applied for long term usage.
K E Y W O R D S
HDAC1, RNAi, antisense DNA
Abbreviation
HDAC1 - Histone deacetylase 1, TNF - Tumor necrosis factor, ssDNA - single strand DNA
I N T R O D U C T I O N
In eukaryotic cells, there are three protein families with classes I, II and III which has multiple interaction pathways. Class I HDACs (HDAC1, 2, 3, 8) are closely related to the transcriptional regulator RPD3 in the yeast S. cerevisiae [1]. Class II HDACs (HDAC4, 5, 6, 7, 9, 10) have protein domains that are similar to HDA1, a deacetylase found in yeast. Class III HDACs form a structurally distinct class of NAD-dependent enzymes. HDACs regulate gene expression by removing the acetyl group from the N-terminal tail of histones. Hypoacetylation of histones decreases the space between a nucleosome and its associated DNA with less accessibility to transcription factors. This leads to transcriptional repression of gene expression [2]. Therefore, traditional HDAC inhibitors can reverse these effects and suppress proliferation of cancer cells and experimental tumors in vivo.
HDAC1 is a class I HDAC which is closely related to various cell responses including the progress of cell cycle, proliferation and gene expression. by regulating RB. HDAC1 inhibitor KBH-A42 decreases the phosphorylation of p38 by dose dependent manner. Also, TNF-alpha, IL-6 and IL-1beta expression is decreased by dose dependent manner.
With the treatment of HDAC1 specific siRNA miR-449a, p27 levels increased via HDAC1 downregulation [3-4]. RNA is folded to have a secondary or tertiary structure by its self-binding in the cytoplasm. Hairpin and loop structures are formed by this mechanism. RNA secondary structure can be identified by bioinformatics approach with generating potential hybrid sites along the mRNA sequence. The energy level and stability differ by the energy levels of the A=U and G=C bonds.
In our study, we used the idea of RNAi and substituted RNA to single strand DNA molecule to see the long-term effects. Previous studies show that interference effects of DNA to a certain mRNA molecule has a highly stable and long term knock down effects in plants. We identified the mRNA secondary structure of HDAC1 comprising the 5’ and 3’ UTR. After all, an antisense DNA single strand (ssDNA) was modeled to mRNA regions where self-hybridization bonding is unlikely or likely to happen. Also, ssDNA that encodes the sequence of miR-449 was also incorporated.
Materials and Methods
I Identification of HDAC1 Sequence
HDAC1 full mRNA sequence was identified and its starting open reading frame (ORF) was also identified. BLAST analysis with HDAC1 mRNA generated statistically similar sequences.
II HDAC1 mRNA Secondary Structure Analysis
Full HDAC1 mRNA sequence in FASTA format was submitted to Wadsworth Bioinformatics Center and generated the possible siRNA binding site with calculated probability. Also, 10 secondary structures of HDAC1 mRNA were generated according to total bond energy and binding energies. Sequences were modeled in circles to identify matching pairs, loop/hairpin connection.
III Generation of Antisense ssDNA Complementary to HDAC1 mRNA
Followed by the secondary structure and siRNA binding probabilities, antisense ssDNA was generated to the sites that have higher probability compared to miR-449 site. The 18-oligomer was generated to gain very high specificity. The number 18 was calculated by the following; four base pair with 18 sequences makes 418 possible configurations, which exceeds the number of 3 billion basepairs, the human genome size. This makes the ssDNA to uniquely bind to the HDAC1 specific site.
Figure 1: The sequence of HDAC1 mRNA The ORF started at the 64th base. The miR-449a was bound to the 1958th base at 3’-UTR
Figure 2: A circle with binding sites was identified according to the self-binding properties of HDAC1. The ensemble centroid was generated for optimization
Figure 3: The siRNA possible sites. Squares indicate the high probability sites and the circles indicate low probability sites
Figure 4: 10 structures according to bond energy. The configuration is sequentially changed according to bonding energy
Tables
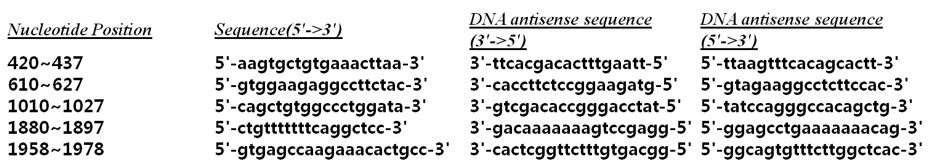
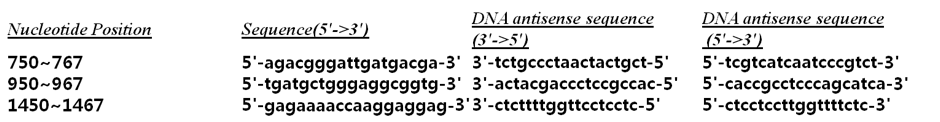
Figure legends
Fig. 1: The site where the probability of siRNA formation was high in HDAC1. The 18oligomer ssDNA was generated to be antisense to this mRNA
Fig. 2: The site where the probability of siRNA formation was relatively low in HDAC1. The 18oligomer ssDNA was generated to be antisense to this mRNA
Results
I HDAC1 Sequence Identification and microRNA Sequence
HDAC1 mRNA was consisted of 2091 nucleotides. The ORF started at the 64th base (Fig. 1). The miR-449a was bound to the 1958th base at 3’-UTR.
II Secondary Structure Hybrid Places and Possible Configuration
The full HDAC1 mRNA sequence was submitted to Sfold 2.2, a software for statistical folding of nucleic acids and studies of regulatory RNAs, provided by Wadwroths Bioinformaticscenter (http://sfold.wadsworth.org). 10 possible mRNA secondary structures with relative energy of -654.1, -646.7, -639.3, -632.1, -624.8, -617.5, -610.2, -602.8, -595.6 and -584.6 (kcal/mol) were generated (Fig. 4). The most stable configuration had an energy level of -687.4 kcal/mol. A circle with binding sites was identified according to the self-binding properties of HDAC1 (Fig. 2).
III Probability of siRNA formation
Each site in mRNA had a different alteration of siRNA binding probability. miR-499a binding site had a probability of 0.90 and there were 4 other sequences with probability higher than 0.90 (420~437 5'-aagtgctgtgaaacttaa-3' / 610~627 5'-gtggaagaggccttctac-3' / 1010~1027 5'-cagctgtggccctggata-3' / 1880~1897 5'-ctgtttttttcaggctcc-3'). Three sites with very low probability were 750~676 (5'-agacgggattgatgacga-3'), 950~967 (5'-tgatgctgggaggcggtg-3') and 1450~1467 (5'-gagaaaaccaaggaggag-3') (Fig. 3).
Discussion
HDAC1 is crucial in epigenetic control of gene expression. We performed HDAC1 mRNA secondary structure analysis by Sfold. There were 10 structures identified with different energetic properties [1]. The hairpin and loop structure were used to identify the possible siRNA targeting sequence. This was actually generated to determine potential ssDNA hybridization sites. As a result, there were 4 positions where the siRNA binding probability was higher than HDAC1 siRNA miR-449a. Three low probability sites were also chosen for positive control.
The previous study performed by HDAC1 siRNA miR-449 had an effect in knock down of HDAC1 and the overexpression of p27 [2]. To demonstrate that ssDNA has a similar effect with microRNA [3], the identical site with antisense DNA incorporated. Other sites with high probability should be tested with experimental procedures in previous reports [5]. The study should be further warranted in comparison of HDAC1 inhibitors and ssDNA in terms of dose dependent and time dependent manners.
Article Info
Article Type
Short CommunicationsPublication history
Received: Wed 28, Mar 2018Accepted: Fri 13, Apr 2018
Published: Mon 30, Apr 2018
Copyright
© 2023 Seung Chan Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.COR.2018.10.008
Author Info
Corresponding Author
Seung Chan KimMedical School & College of Medicine, Yonsei University, Seoul, Republic of Korea
Figures & Tables
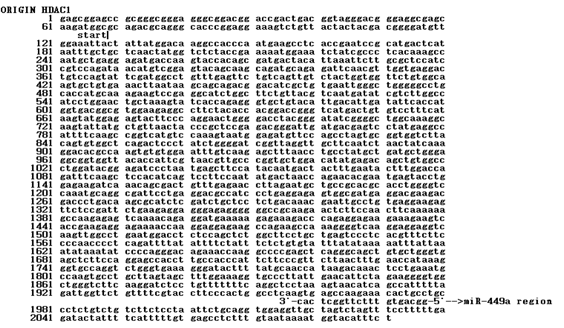
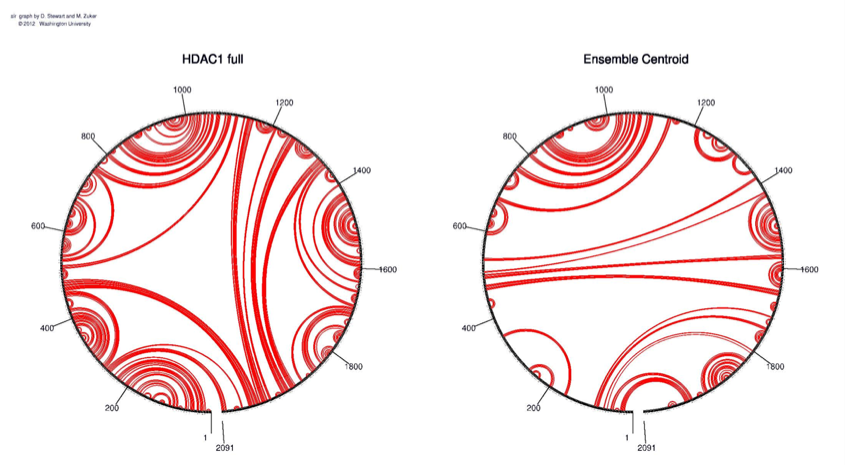


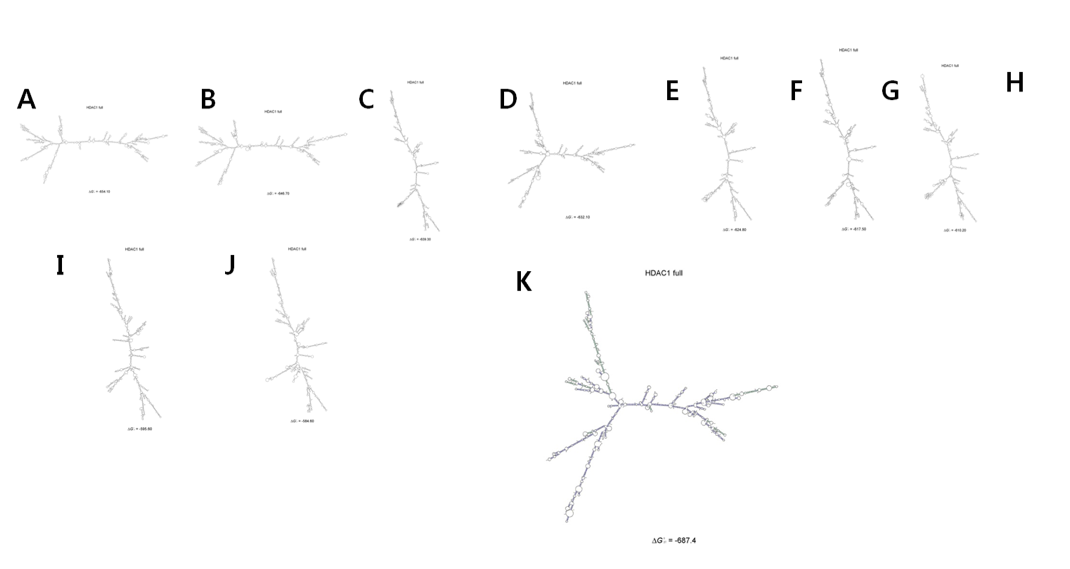
Figure legends
Fig. 1: The sequence of HDAC1 mRNA The ORF started at the 64th base. The miR-449a was bound to the 1958th base at 3’-UTR
Fig. 2: A circle with binding sites was identified according to the self-binding properties of HDAC1. The ensemble centroid was generated for optimization
Fig. 3: The siRNA possible sites. Squares indicate the high probability sites and the circles indicate low probability sites
Fig. 4: 10 structures according to bond energy. The configuration is sequentially changed according to bonding energy


Figure legends
Fig. 1: The site where the probability of siRNA formation was high in HDAC1. The 18oligomer ssDNA was generated to be antisense to this mRNA
Fig. 2: The site where the probability of siRNA formation was relatively low in HDAC1. The 18oligomer ssDNA was generated to be antisense to this mRNA
References
1. Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, et al. (2009) miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 28: 1714-1724. [Crossref]
2. Choi Y, Park SK, Kim HM, Kang JS, Yoon YD, et al. (2008) Histone deacetylase inhibitor KBH-A42 inhibits cytokine production in RAW 264.7 macrophage cells and in vivo endotoxemia model. Exp Mol Med 40: 574-581. [Crossref]
3. Sen A, Nelson TJ, Alkon DL (2015) ApoE4 and Aβ Oligomers Reduce BDNF Expression via HDAC Nuclear Translocation. J Neurosci 35: 7538-7551. [Crossref]
4. Xiao JJ, Huang Y, Dai Z, Sadée W, Chen J, et al. (2005) Chemoresistance to depsipeptide FK228 [(E)-(1S,4S,10S,21R)-7-[(Z)-ethylidene]-4,21-diisopropyl-2-oxa-12,13-dithia-5,8,2,0,23-tetraazabicyclo[8,7,6]-tricos-16-ene-3,6,9,22-pentanone] is mediated by reversible MDR1 induction in human cancer cell lines. J Pharmacol Exp Ther 314: 467-475. [Crossref]
5. Xiao JJ, Foraker AB, Swaan PW, Liu S, Huang Y, et al. (2005) Efflux of depsipeptide FK228 (FR901228, NSC-630176) is mediated by P-glycoprotein and multidrug resistance-associated protein 1. J Pharmacol Exp Ther 313: 268-276. [Crossref]
